A Foxtail mosaic virus Vector for Virus-Induced Gene Silencing in Maize
- PMID: 27208311
- PMCID: PMC4902600
- DOI: 10.1104/pp.16.00172
A Foxtail mosaic virus Vector for Virus-Induced Gene Silencing in Maize
Abstract
Plant viruses have been widely used as vectors for foreign gene expression and virus-induced gene silencing (VIGS). A limited number of viruses have been developed into viral vectors for the purposes of gene expression or VIGS in monocotyledonous plants, and among these, the tripartite viruses Brome mosaic virus and Cucumber mosaic virus have been shown to induce VIGS in maize (Zea mays). We describe here a new DNA-based VIGS system derived from Foxtail mosaic virus (FoMV), a monopartite virus that is able to establish systemic infection and silencing of endogenous maize genes homologous to gene fragments inserted into the FoMV genome. To demonstrate VIGS applications of this FoMV vector system, four genes, phytoene desaturase (functions in carotenoid biosynthesis), lesion mimic22 (encodes a key enzyme of the porphyrin pathway), iojap (functions in plastid development), and brown midrib3 (caffeic acid O-methyltransferase), were silenced and characterized in the sweet corn line Golden × Bantam. Furthermore, we demonstrate that the FoMV infectious clone establishes systemic infection in maize inbred lines, sorghum (Sorghum bicolor), and green foxtail (Setaria viridis), indicating the potential wide applications of this viral vector system for functional genomics studies in maize and other monocots.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures
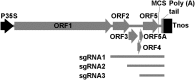

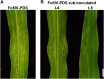
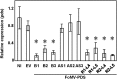
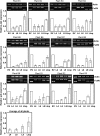

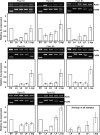

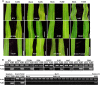
Similar articles
-
Virus-Induced Gene Silencing in Maize with a Foxtail mosaic virus Vector.Methods Mol Biol. 2018;1676:129-139. doi: 10.1007/978-1-4939-7315-6_7. Methods Mol Biol. 2018. PMID: 28986907
-
Foxtail Mosaic Virus-Induced Gene Silencing in Monocot Plants.Plant Physiol. 2016 Jul;171(3):1801-7. doi: 10.1104/pp.16.00010. Epub 2016 May 25. Plant Physiol. 2016. PMID: 27225900 Free PMC article.
-
Direct Agroinoculation of Maize Seedlings by Injection with Recombinant Foxtail Mosaic Virus and Sugarcane Mosaic Virus Infectious Clones.J Vis Exp. 2021 Feb 27;(168). doi: 10.3791/62277. J Vis Exp. 2021. PMID: 33720142
-
Foxtail mosaic virus: A tool for gene function analysis in maize and other monocots.Mol Plant Pathol. 2023 Jul;24(7):811-822. doi: 10.1111/mpp.13330. Epub 2023 Apr 10. Mol Plant Pathol. 2023. PMID: 37036421 Free PMC article. Review.
-
Gene silencing approaches through virus-based vectors: speeding up functional genomics in monocots.Plant Mol Biol. 2019 May;100(1-2):3-18. doi: 10.1007/s11103-019-00854-6. Epub 2019 Mar 8. Plant Mol Biol. 2019. PMID: 30850930 Review.
Cited by
-
Engineering Maize rayado fino virus for virus-induced gene silencing.Plant Direct. 2020 Aug 6;4(8):e00224. doi: 10.1002/pld3.224. eCollection 2020 Aug. Plant Direct. 2020. PMID: 32783020 Free PMC article.
-
A cassava common mosaic virus vector for virus-induced gene silencing in cassava.Plant Methods. 2021 Jul 12;17(1):74. doi: 10.1186/s13007-021-00775-w. Plant Methods. 2021. PMID: 34247636 Free PMC article.
-
Plant Virology Delivers Diverse Toolsets for Biotechnology.Viruses. 2020 Nov 23;12(11):1338. doi: 10.3390/v12111338. Viruses. 2020. PMID: 33238421 Free PMC article. Review.
-
RNA Interference: Promising Approach to Combat Plant Viruses.Int J Mol Sci. 2022 May 10;23(10):5312. doi: 10.3390/ijms23105312. Int J Mol Sci. 2022. PMID: 35628126 Free PMC article. Review.
-
Virus-induced gene silencing for in planta validation of gene function in cucurbits.Plant Physiol. 2022 Nov 28;190(4):2366-2379. doi: 10.1093/plphys/kiac363. Plant Physiol. 2022. PMID: 35944218 Free PMC article.
References
-
- Avesani L, Marconi G, Morandini F, Albertini E, Bruschetta M, Bortesi L, Pezzotti M, Porceddu A (2007) Stability of Potato virus X expression vectors is related to insert size: implications for replication models and risk assessment. Transgenic Res 16: 587–597 - PubMed
-
- Bancroft JB, Rouleau M, Johnston R, Prins L, Mackie GA (1991) The entire nucleotide sequence of foxtail mosaic virus RNA. J Gen Virol 72: 2173–2181 - PubMed
-
- Barrière Y, Argillier O (1993) Brown-midrib genes of maize: a review. Agronomie 13: 865–876
-
- Becker A. (2013) Virus-induced gene silencing: methods and protocols. Methods Mol Biol 975: 221
-
- Benavente LM, Ding XS, Redinbaugh MG, Nelson RS, Balint-Kurti P (2012) Virus-induced gene silencing in diverse maize lines using the Brome mosaic virus-based silencing vector. Maydica 57: 206–214
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

