The piRNA pathway is developmentally regulated during spermatogenesis in Drosophila
- PMID: 27208314
- PMCID: PMC4911912
- DOI: 10.1261/rna.055996.116
The piRNA pathway is developmentally regulated during spermatogenesis in Drosophila
Abstract
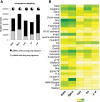
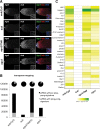
PIWI-interacting RNAs (piRNAs) are predominantly produced in animal gonads to suppress transposons during germline development. Our understanding about the piRNA biogenesis and function is predominantly from studies of the Drosophila female germline. piRNA pathway function in the male germline, however, remains poorly understood. To study overall and stage-specific features of piRNAs during spermatogenesis, we analyzed small RNAs extracted from entire wild-type testes and stage-specific arrest mutant testes enriched with spermatogonia or primary spermatocytes. We show that most active piRNA clusters in the female germline do not majorly contribute to piRNAs in testes, and abundance patterns of piRNAs mapping to different transposon families also differ between male and female germlines. piRNA production is regulated in a stage-specific manner during spermatogenesis. The piRNAs in spermatogonia-enriched testes are predominantly transposon-mapping piRNAs, and almost half of those exhibit a ping-pong signature. In contrast, the primary spermatocyte-enriched testes have a dramatically high amount of piRNAs targeting repeats like suppressor of stellate and AT-chX The transposon-mapping piRNAs in the primary spermatocyte stages lacking Argonaute3 expression also show a ping-pong signature, albeit to a lesser extent. Consistently, argonaute3 mutant testes also retain ping-pong signature-bearing piRNAs, suggesting that a noncanonical ping-pong cycle might act during spermatogenesis. Our study shows stage-specific regulation of piRNA biogenesis during spermatogenesis: An active ping-pong cycle produces abundant transposon-mapping piRNAs in spermatogonia, while in primary spermatocytes, piRNAs act to suppress the repeats and transposons.
Keywords: Drosophila; PIWI proteins; RNA silencing; germline; piRNA; spermatogenesis.
© 2016 Quénerch'du et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027. - PubMed
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. - PubMed
-
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
