Reassortment in segmented RNA viruses: mechanisms and outcomes
- PMID: 27211789
- PMCID: PMC5119462
- DOI: 10.1038/nrmicro.2016.46
Reassortment in segmented RNA viruses: mechanisms and outcomes
Abstract
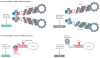
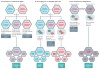
Segmented RNA viruses are widespread in nature and include important human, animal and plant pathogens, such as influenza viruses and rotaviruses. Although the origin of RNA virus genome segmentation remains elusive, a major consequence of this genome structure is the capacity for reassortment to occur during co-infection, whereby segments are exchanged among different viral strains. Therefore, reassortment can create viral progeny that contain genes that are derived from more than one parent, potentially conferring important fitness advantages or disadvantages to the progeny virus. However, for segmented RNA viruses that package their multiple genome segments into a single virion particle, reassortment also requires genetic compatibility between parental strains, which occurs in the form of conserved packaging signals, and the maintenance of RNA and protein interactions. In this Review, we discuss recent studies that examined the mechanisms and outcomes of reassortment for three well-studied viral families - Cystoviridae, Orthomyxoviridae and Reoviridae - and discuss how these findings provide new perspectives on the replication and evolution of segmented RNA viruses.
Conflict of interest statement
statement The authors declare no competing interests.
Figures


References
-
- Tate JE, et al. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:136–141. - PubMed
-
- Maclachlan NJ, Mayo CE. Potential strategies for control of bluetongue, a globally emerging, Culicoides-transmitted viral disease of ruminant livestock and wildlife. Antiviral Res. 2013;99:79–90. - PubMed
-
- Mahgoub HA, Bailey M, Kaiser P. An overview of infectious bursal disease. Arch Virol. 2012;157:2047–2057. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

