Functional interaction of histone deacetylase 5 (HDAC5) and lysine-specific demethylase 1 (LSD1) promotes breast cancer progression
- PMID: 27212032
- PMCID: PMC5121103
- DOI: 10.1038/onc.2016.186
Functional interaction of histone deacetylase 5 (HDAC5) and lysine-specific demethylase 1 (LSD1) promotes breast cancer progression
Abstract
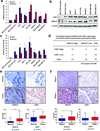
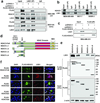
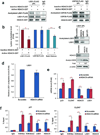
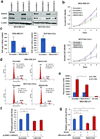
We have previously demonstrated that crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases (HDACs) facilitates breast cancer proliferation. However, the underlying mechanisms are largely unknown. Here, we report that expression of HDAC5 and LSD1 proteins were positively correlated in human breast cancer cell lines and tissue specimens of primary breast tumors. Protein expression of HDAC5 and LSD1 was significantly increased in primary breast cancer specimens in comparison with matched-normal adjacent tissues. Using HDAC5 deletion mutants and co-immunoprecipitation studies, we showed that HDAC5 physically interacted with the LSD1 complex through its domain containing nuclear localization sequence and phosphorylation sites. Although the in vitro acetylation assays revealed that HDAC5 decreased LSD1 protein acetylation, small interfering RNA (siRNA)-mediated HDAC5 knockdown did not alter the acetylation level of LSD1 in MDA-MB-231 cells. Overexpression of HDAC5 stabilized LSD1 protein and decreased the nuclear level of H3K4me1/me2 in MDA-MB-231 cells, whereas loss of HDAC5 by siRNA diminished LSD1 protein stability and demethylation activity. We further demonstrated that HDAC5 promoted the protein stability of USP28, a bona fide deubiquitinase of LSD1. Overexpression of USP28 largely reversed HDAC5-KD-induced LSD1 protein degradation, suggesting a role of HDAC5 as a positive regulator of LSD1 through upregulation of USP28 protein. Depletion of HDAC5 by shRNA hindered cellular proliferation, induced G1 cell cycle arrest, and attenuated migration and colony formation of breast cancer cells. A rescue study showed that increased growth of MDA-MB-231 cells by HDAC5 overexpression was reversed by concurrent LSD1 depletion, indicating that tumor-promoting activity of HDAC5 is an LSD1 dependent function. Moreover, overexpression of HDAC5 accelerated cellular proliferation and promoted acridine mutagen ICR191-induced transformation of MCF10A cells. Taken together, these results suggest that HDAC5 is critical in regulating LSD1 protein stability through post-translational modification, and the HDAC5-LSD1 axis has an important role in promoting breast cancer development and progression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
HDAC5-LSD1 axis regulates antineoplastic effect of natural HDAC inhibitor sulforaphane in human breast cancer cells.Int J Cancer. 2018 Sep 15;143(6):1388-1401. doi: 10.1002/ijc.31419. Epub 2018 Apr 20. Int J Cancer. 2018. PMID: 29633255 Free PMC article.
-
Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells.Carcinogenesis. 2013 Jun;34(6):1196-207. doi: 10.1093/carcin/bgt033. Epub 2013 Jan 25. Carcinogenesis. 2013. PMID: 23354309 Free PMC article.
-
The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells.Cell Rep. 2013 Oct 17;5(1):224-36. doi: 10.1016/j.celrep.2013.08.030. Epub 2013 Sep 26. Cell Rep. 2013. PMID: 24075993 Free PMC article.
-
LSD1: more than demethylation of histone lysine residues.Exp Mol Med. 2020 Dec;52(12):1936-1947. doi: 10.1038/s12276-020-00542-2. Epub 2020 Dec 14. Exp Mol Med. 2020. PMID: 33318631 Free PMC article. Review.
-
Biological roles of LSD1 beyond its demethylase activity.Cell Mol Life Sci. 2020 Sep;77(17):3341-3350. doi: 10.1007/s00018-020-03489-9. Epub 2020 Mar 19. Cell Mol Life Sci. 2020. PMID: 32193608 Free PMC article. Review.
Cited by
-
Neutralizing negative epigenetic regulation by HDAC5 enhances human haematopoietic stem cell homing and engraftment.Nat Commun. 2018 Jul 16;9(1):2741. doi: 10.1038/s41467-018-05178-5. Nat Commun. 2018. PMID: 30013077 Free PMC article.
-
HDAC5-mediated deacetylation and nuclear localisation of SOX9 is critical for tamoxifen resistance in breast cancer.Br J Cancer. 2019 Dec;121(12):1039-1049. doi: 10.1038/s41416-019-0625-0. Epub 2019 Nov 6. Br J Cancer. 2019. PMID: 31690832 Free PMC article.
-
USP28: Oncogene or Tumor Suppressor? A Unifying Paradigm for Squamous Cell Carcinoma.Cells. 2021 Oct 4;10(10):2652. doi: 10.3390/cells10102652. Cells. 2021. PMID: 34685632 Free PMC article. Review.
-
HDAC5-LSD1 axis regulates antineoplastic effect of natural HDAC inhibitor sulforaphane in human breast cancer cells.Int J Cancer. 2018 Sep 15;143(6):1388-1401. doi: 10.1002/ijc.31419. Epub 2018 Apr 20. Int J Cancer. 2018. PMID: 29633255 Free PMC article.
-
Lysine-specific demethylase 1 (LSD1) destabilizes p62 and inhibits autophagy in gynecologic malignancies.Oncotarget. 2017 Aug 10;8(43):74434-74450. doi: 10.18632/oncotarget.20158. eCollection 2017 Sep 26. Oncotarget. 2017. PMID: 29088798 Free PMC article.
References
-
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. - PubMed
-
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–435. - PubMed
-
- Lee M, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–435. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

