Root Regeneration Triggers an Embryo-like Sequence Guided by Hormonal Interactions
- PMID: 27212234
- PMCID: PMC4912400
- DOI: 10.1016/j.cell.2016.04.046
Root Regeneration Triggers an Embryo-like Sequence Guided by Hormonal Interactions
Abstract
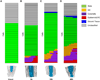
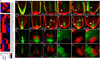
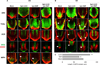
Plant roots can regenerate after excision of their tip, including the stem cell niche. To determine which developmental program mediates such repair, we applied a combination of lineage tracing, single-cell RNA sequencing, and marker analysis to test different models of tissue reassembly. We show that multiple cell types can reconstitute stem cells, demonstrating the latent potential of untreated plant cells. The transcriptome of regenerating cells prior to stem cell activation resembles that of an embryonic root progenitor. Regeneration defects are more severe in embryonic than in adult root mutants. Furthermore, the signaling domains of the hormones auxin and cytokinin mirror their embryonic dynamics and manipulation of both hormones alters the position of new tissues and stem cell niche markers. Our findings suggest that plant root regeneration follows, on a larger scale, the developmental stages of embryonic patterning and is guided by spatial information provided by complementary hormone domains.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




References
-
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:119–120. - PubMed
-
- Alvarado AS, Tsonis Pa. Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 2006;7:873–884. - PubMed
-
- Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009;57:626–644. - PubMed
-
- Bellini C, Pacurar DI, Perrone I, Bellini Catherine, Pacurar Daniel I, Perrone Irene. Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 2014;65:639–666. - PubMed
-
- Bennett T, Scheres B. Root development-two meristems for the price of one? Curr. Top. Dev. Biol. 2010;91:67–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

