Coexisting Liquid Phases Underlie Nucleolar Subcompartments
- PMID: 27212236
- PMCID: PMC5127388
- DOI: 10.1016/j.cell.2016.04.047
Coexisting Liquid Phases Underlie Nucleolar Subcompartments
Abstract
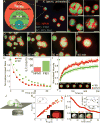
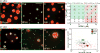
The nucleolus and other ribonucleoprotein (RNP) bodies are membrane-less organelles that appear to assemble through phase separation of their molecular components. However, many such RNP bodies contain internal subcompartments, and the mechanism of their formation remains unclear. Here, we combine in vivo and in vitro studies, together with computational modeling, to show that subcompartments within the nucleolus represent distinct, coexisting liquid phases. Consistent with their in vivo immiscibility, purified nucleolar proteins phase separate into droplets containing distinct non-coalescing phases that are remarkably similar to nucleoli in vivo. This layered droplet organization is caused by differences in the biophysical properties of the phases-particularly droplet surface tension-which arises from sequence-encoded features of their macromolecular components. These results suggest that phase separation can give rise to multilayered liquids that may facilitate sequential RNA processing reactions in a variety of RNP bodies. PAPERCLIP.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




References
-
- AARTS DG, SCHMIDT M, LEKKERKERKER HN. Direct visual observation of thermal capillary waves. Science. 2004;304:847–850. - PubMed
-
- BOISVERT FM, VAN KONINGSBRUGGEN S, NAVASCUES J, LAMOND AI. The multifunctional nucleolus. Nat Rev Mol Cell Bio. 2007;8:574–585. - PubMed
-
- BRANGWYNNE CP, ECKMANN CR, COURSON DS, RYBARSKA A, HOEGE C, GHARAKHANI J, JULICHER F, HYMAN AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

