Relative rate and location of intra-host HIV evolution to evade cellular immunity are predictable
- PMID: 27212475
- PMCID: PMC4879252
- DOI: 10.1038/ncomms11660
Relative rate and location of intra-host HIV evolution to evade cellular immunity are predictable
Abstract
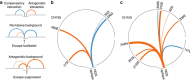
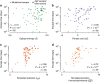
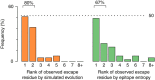
Human immunodeficiency virus (HIV) evolves within infected persons to escape being destroyed by the host immune system, thereby preventing effective immune control of infection. Here, we combine methods from evolutionary dynamics and statistical physics to simulate in vivo HIV sequence evolution, predicting the relative rate of escape and the location of escape mutations in response to T-cell-mediated immune pressure in a cohort of 17 persons with acute HIV infection. Predicted and clinically observed times to escape immune responses agree well, and we show that the mutational pathways to escape depend on the viral sequence background due to epistatic interactions. The ability to predict escape pathways and the duration over which control is maintained by specific immune responses open the door to rational design of immunotherapeutic strategies that might enable long-term control of HIV infection. Our approach enables intra-host evolution of a human pathogen to be predicted in a probabilistic framework.
Figures
References
-
- Phillips R. E. et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354, 453–459 (1991). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

