A gene expression microarray for Nicotiana benthamiana based on de novo transcriptome sequence assembly
- PMID: 27213006
- PMCID: PMC4875705
- DOI: 10.1186/s13007-016-0128-4
A gene expression microarray for Nicotiana benthamiana based on de novo transcriptome sequence assembly
Abstract
Background: Nicotiana benthamiana has been widely used in laboratories around the world for studying plant-pathogen interactions and posttranscriptional gene expression silencing. Yet the exploration of its transcriptome has lagged behind due to the lack of both adequate sequence information and genome-wide analysis tools, such as DNA microarrays. Despite the increasing use of high-throughput sequencing technologies, the DNA microarrays still remain a popular gene expression tool, because they are cheaper and less demanding regarding bioinformatics skills and computational effort.
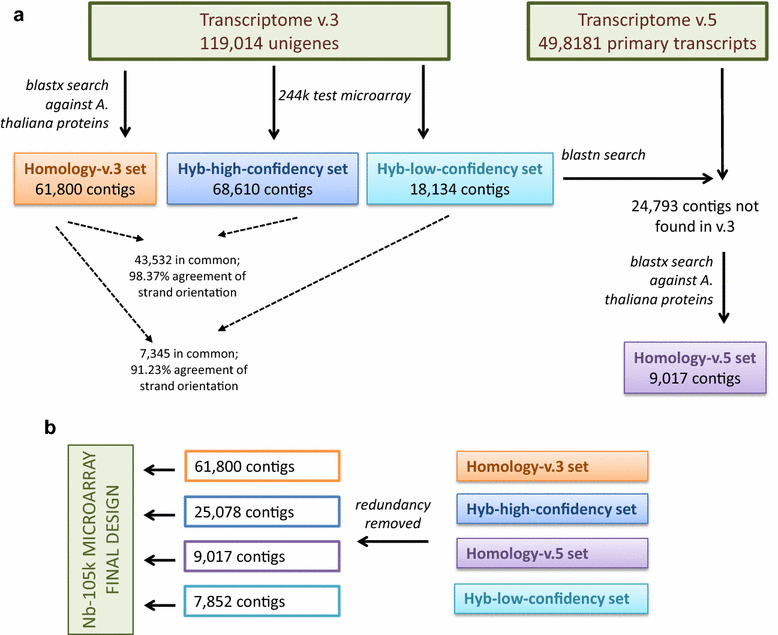
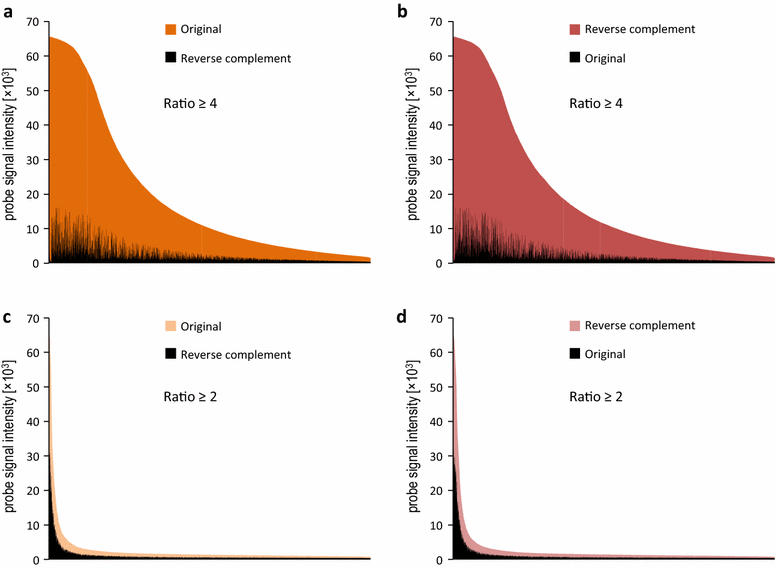
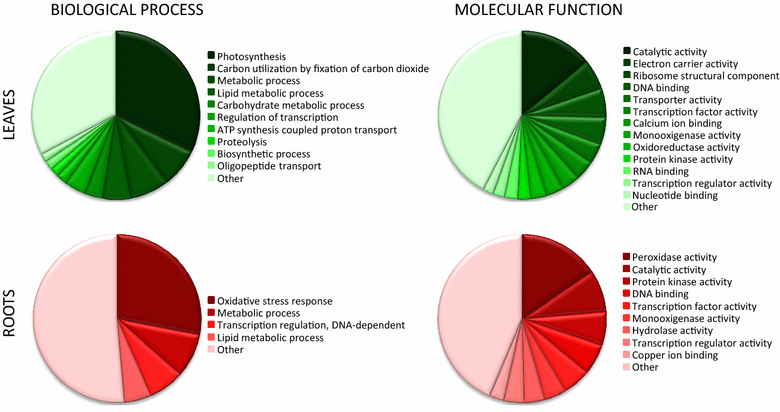
Results: We designed a gene expression microarray with 103,747 60-mer probes, based on two recently published versions of N. benthamiana transcriptome (v.3 and v.5). Both versions were reconstructed from RNA-Seq data of non-strand-specific pooled-tissue libraries, so we defined the sense strand of the contigs prior to designing the probe. To accomplish this, we combined a homology search against Arabidopsis thaliana proteins and hybridization to a test 244k microarray containing pairs of probes, which represented individual contigs. We identified the sense strand in 106,684 transcriptome contigs and used this information to design an Nb-105k microarray on an Agilent eArray platform. Following hybridization of RNA samples from N. benthamiana roots and leaves we demonstrated that the new microarray had high specificity and sensitivity for detection of differentially expressed transcripts. We also showed that the data generated with the Nb-105k microarray may be used to identify incorrectly assembled contigs in the v.5 transcriptome, by detecting inconsistency in the gene expression profiles, which is indicated using multiple microarray probes that match the same v.5 primary transcripts.
Conclusions: We provided a complete design of an oligonucleotide microarray that may be applied to the research of N. benthamiana transcriptome. This, in turn, will allow the N. benthamiana research community to take full advantage of microarray capabilities for studying gene expression in this plant. Additionally, by defining the sense orientation of over 106,000 contigs, we substantially improved the functional information on the N. benthamiana transcriptome. The simple hybridization-based approach for detecting the sense orientation of computationally assembled sequences can be used for updating the transcriptomes of other non-model organisms, including cases where no significant homology to known proteins exists.
Keywords: Coding strand; Leaf and root transcriptome; Microarray; Nicotiana benthamiana.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

