Origin and evolution of the T cell repertoire after posttransplantation cyclophosphamide
- PMID: 27213183
- PMCID: PMC4874509
- DOI: 10.1172/jci.insight.86252
Origin and evolution of the T cell repertoire after posttransplantation cyclophosphamide
Abstract
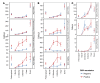
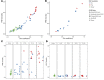
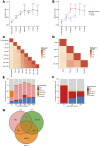
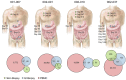
Posttransplantation cyclophosphamide (PTCy) effectively prevents graft-versus-host disease (GVHD), but its immunologic impact is poorly understood. We assessed lymphocyte reconstitution via flow cytometry (n = 74) and antigen receptor sequencing (n = 35) in recipients of myeloablative, HLA-matched allogeneic BM transplantation using PTCy. Recovering T cells were primarily phenotypically effector memory with lower T cell receptor β (TRB) repertoire diversity than input donor repertoires. Recovering B cells were predominantly naive with immunoglobulin heavy chain locus (IGH) repertoire diversity similar to donors. Numerical T cell reconstitution and TRB diversity were strongly associated with recipient cytomegalovirus seropositivity. Global similarity between input donor and recipient posttransplant repertoires was uniformly low at 1-2 months after transplant but increased over the balance of the first posttransplant year. Blood TRB repertoires at ≥3 months after transplant were often dominated by clones present in the donor blood/marrow memory CD8+ compartment. Limited overlap was observed between the TRB repertoires of T cells infiltrating the skin or gastrointestinal tract versus the blood. Although public TRB sequences associated with herpesvirus- or alloantigen-specific CD8+ T cells were detected in some patients, posttransplant TRB and IGH repertoires were unique to each individual. These data define the immune dynamics occurring after PTCy and establish a benchmark against which immune recovery after other transplantation approaches can be compared.
Figures



References
-
- Bashey A, et al. T-cell-replete HLA-haploidentical hematopoietic transplantation for hematologic malignancies using post-transplantation cyclophosphamide results in outcomes equivalent to those of contemporaneous HLA-matched related and unrelated donor transplantation. J Clin Oncol. 2013;31(10):1310–1316. doi: 10.1200/JCO.2012.44.3523. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

