Characterization and Functional Analysis of 4-Coumarate:CoA Ligase Genes in Mul-berry
- PMID: 27213624
- PMCID: PMC4877003
- DOI: 10.1371/journal.pone.0155814
Characterization and Functional Analysis of 4-Coumarate:CoA Ligase Genes in Mul-berry
Erratum in
-
Correction: Characterization and Functional Analysis of 4-Coumarate:CoA Ligase Genes in Mulberry.PLoS One. 2016 Jun 8;11(6):e0157414. doi: 10.1371/journal.pone.0157414. eCollection 2016. PLoS One. 2016. PMID: 27276057 Free PMC article.
Abstract
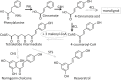
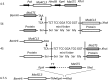
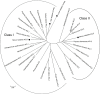
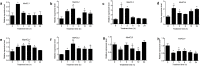
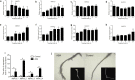
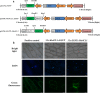
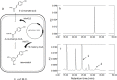
A small, multigene family encodes 4-coumarate:CoA ligases (4CLs) that catalyze the ligation of CoA to hydroxycinnamic acids, a branch point directing metabolites to flavonoid or monolignol pathways. In this study, we characterized four 4CL genes from M. notabilis Genome Database, and cloned four Ma4CL genes from M. atropurpurea cv. Jialing No.40. A tissue-specific expression analysis indicated that Ma4CL3 was expressed at higher levels than the other genes, and that Ma4CL3 was strongly expressed in root bark, stem bark, and old leaves. Additionally, the expression pattern of Ma4CL3 was similar to the trend of the total flavonoid content throughout fruit development. A phylogenetic analysis suggested that Mn4CL1, Mn4CL2, and Mn4CL4 belong to class I 4CLs, and Mn4CL3 belongs to class II 4CLs. Ma4CL genes responded differently to a series of stresses. Ma4CL3 expression was higher than that of the other Ma4CL genes following wounding, salicylic acid, and ultraviolet treatments. An in vitro enzyme assay indicated that 4-coumarate acid was the best substrate among cinnamic acid, 4-coumarate acid, and caffeate acid, but no catalytic activity to sinapate acid and ferulate acid. The results of subcellular localization experiments showed that Ma4CL3 localized to the cytomembrane, where it activated transcription. We used different vectors and strategies to fuse Ma4CL3 with stilbene synthase (STS) to construct four Ma4CL-MaSTS co-expression systems to generate resveratrol. The results indicated that only a transcriptional fusion vector, pET-Ma4CL3-T-MaSTS, which utilized a T7 promoter and lac operator for the expression of MaSTS, could synthesize resveratrol.
Conflict of interest statement
Figures




Similar articles
-
4-coumarate: CoA ligase partitions metabolites for eugenol biosynthesis.Plant Cell Physiol. 2013 Aug;54(8):1238-52. doi: 10.1093/pcp/pct073. Epub 2013 May 14. Plant Cell Physiol. 2013. PMID: 23677922
-
Characterization of Stilbene Synthase Genes in Mulberry (Morus atropurpurea) and Metabolic Engineering for the Production of Resveratrol in Escherichia coli.J Agric Food Chem. 2017 Mar 1;65(8):1659-1668. doi: 10.1021/acs.jafc.6b05212. Epub 2017 Feb 14. J Agric Food Chem. 2017. PMID: 28168876
-
Ectopic expression of a loblolly pine class II 4-coumarate:CoA ligase alters soluble phenylpropanoid metabolism but not lignin biosynthesis in Populus.Plant Cell Physiol. 2014 Sep;55(9):1669-78. doi: 10.1093/pcp/pcu098. Epub 2014 Jul 12. Plant Cell Physiol. 2014. PMID: 25016610
-
Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation.Phytochemistry. 2005 Sep;66(17):2072-91. doi: 10.1016/j.phytochem.2005.06.022. Phytochemistry. 2005. PMID: 16099486
-
Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants.Planta. 2018 Nov;248(5):1063-1078. doi: 10.1007/s00425-018-2965-z. Epub 2018 Aug 4. Planta. 2018. PMID: 30078075 Review.
Cited by
-
Flavonoids Accumulation in Fruit Peel and Expression Profiling of Related Genes in Purple (Passiflora edulis f. edulis) and Yellow (Passiflora edulis f. flavicarpa) Passion Fruits.Plants (Basel). 2021 Oct 20;10(11):2240. doi: 10.3390/plants10112240. Plants (Basel). 2021. PMID: 34834602 Free PMC article.
-
Revealing floral metabolite network in tuberose that underpins scent volatiles synthesis, storage and emission.Plant Mol Biol. 2021 Aug;106(6):533-554. doi: 10.1007/s11103-021-01171-7. Epub 2021 Jul 14. Plant Mol Biol. 2021. PMID: 34263437
-
Pluronic F-68 Improves Callus Proliferation of Recalcitrant Rice Cultivar via Enhanced Carbon and Nitrogen Metabolism and Nutrients Uptake.Front Plant Sci. 2021 Jun 2;12:667434. doi: 10.3389/fpls.2021.667434. eCollection 2021. Front Plant Sci. 2021. PMID: 34149763 Free PMC article.
-
Combined Metabolome and Transcriptome Analyses Reveal the Effects of Mycorrhizal Fungus Ceratobasidium sp. AR2 on the Flavonoid Accumulation in Anoectochilus roxburghii during Different Growth Stages.Int J Mol Sci. 2020 Jan 15;21(2):564. doi: 10.3390/ijms21020564. Int J Mol Sci. 2020. PMID: 31952330 Free PMC article.
-
Synergistic potential of teriflunomide with fluconazole against resistant Candida albicans in vitro and in vivo.Front Cell Infect Microbiol. 2023 Dec 19;13:1282320. doi: 10.3389/fcimb.2023.1282320. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38169891 Free PMC article.
References
-
- Beggs C and Wellman E (1985) Analysis of light-controlled anthocyanin formation in coleoptiles of Zea mays L.: the role of UV-B, blue, red and far-red light. Photochem Photobiol. 41:481–486.
-
- Chappell J and Hahlbrock K (1984) Transcription of plant defence genes in response to UV light or fungal elicitor. Nature. 311:76–78.
-
- Hahlbrock K and Scheel D (1989) Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 40:347–369.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

