Detection of Severe Acute Respiratory Syndrome-Like, Middle East Respiratory Syndrome-Like Bat Coronaviruses and Group H Rotavirus in Faeces of Korean Bats
- PMID: 27213718
- PMCID: PMC7169817
- DOI: 10.1111/tbed.12515
Detection of Severe Acute Respiratory Syndrome-Like, Middle East Respiratory Syndrome-Like Bat Coronaviruses and Group H Rotavirus in Faeces of Korean Bats
Abstract
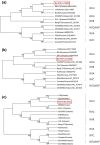
Bat species around the world have recently been recognized as major reservoirs of several zoonotic viruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), Nipah virus and Hendra virus. In this study, consensus primer-based reverse transcriptase polymerase chain reactions (RT-PCRs) and high-throughput sequencing were performed to investigate viruses in bat faecal samples collected at 11 natural bat habitat sites from July to December 2015 in Korea. Diverse coronaviruses were first detected in Korean bat faeces, including alphacoronaviruses, SARS-CoV-like and MERS-CoV-like betacoronaviruses. In addition, we identified a novel bat rotavirus belonging to group H rotavirus which has only been described in human and pigs until now. Therefore, our results suggest the need for continuing surveillance and additional virological studies in domestic bat.
Keywords: Korea; Middle East respiratory syndrome; bat; coronavirus; group H rotavirus; severe acute respiratory syndrome.
© 2016 Blackwell Verlag GmbH.
Figures

References
-
- Alam, M. M. , Kobayashi N., Ishino M., Ahmed M. S., Ahmed M. U., Paul S. K., Muzumdar B. K., Hussain Z., Wang Y. H., and Naik T. N., 2007: Genetic analysis of an ADRV‐N‐like novel rotavirus strain B219 detected in a sporadic case of adult diarrhea in Bangladesh. Arch. Virol. 152, 199–208. - PubMed
-
- Annan, A. , Baldwin H. J., Corman V. M., Klose S. M., Owusu M., Nkrumah E. E., Badu E. K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y. A., Kalko E. K. V., Lina P. H. C., Godlevska E. V., Reusken C., Seebens A., Gloza‐Rausch F., Vallo P., Tschapka M., Drosten C., and Drexler J. F., 2013: Human betacoronavirus 2c EMC/2012–related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 19, 456–459. - PMC - PubMed
-
- Chu, D. K. W. , Peiris J. S. M., Chen H., Guan Y., and Poon L. L. M., 2008: Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co‐infections in Miniopterus bats. J. Gen. Virol. 89, 1282–1287. - PubMed
-
- Esona, M. D. , Mijatovic‐Rustempasic S., Conrardy C., Tong S., Kuzmin I. V., Agwanda B., Breiman R. F., Banyai K., Niezgoda M., Rupprecht C. E., Gentsch J. R., and Bowen M. D., 2010: Reassortant group A rotavirus from straw‐colored fruit bat (Eidolon helvum). Emerg. Infect. Dis. 16, 1844–1852. - PMC - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

