Comparison of Cas9 activators in multiple species
- PMID: 27214048
- PMCID: PMC4927356
- DOI: 10.1038/nmeth.3871
Comparison of Cas9 activators in multiple species
Abstract
Several programmable transcription factors exist based on the versatile Cas9 protein, yet their relative potency and effectiveness across various cell types and species remain unexplored. Here, we compare Cas9 activator systems and examine their ability to induce robust gene expression in several human, mouse, and fly cell lines. We also explore the potential for improved activation through the combination of the most potent activator systems, and we assess the role of cooperativity in maximizing gene expression.
Figures

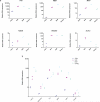
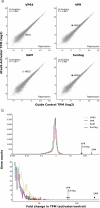
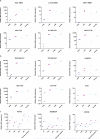
References
-
- Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

