Transcriptomic and proteomic analyses of core metabolism in Clostridium termitidis CT1112 during growth on α-cellulose, xylan, cellobiose and xylose
- PMID: 27215540
- PMCID: PMC4877739
- DOI: 10.1186/s12866-016-0711-x
Transcriptomic and proteomic analyses of core metabolism in Clostridium termitidis CT1112 during growth on α-cellulose, xylan, cellobiose and xylose
Abstract
Background: Clostridium termitidis CT1112 is an anaerobic, Gram-positive, mesophilic, spore-forming, cellulolytic bacterium, originally isolated from the gut of a wood feeding termite Nasusitermes lujae. It has the ability to hydrolyze both cellulose and hemicellulose, and ferment the degradation products to acetate, formate, ethanol, lactate, H2, and CO2. It is therefore ges in gene and gene product expression during growth of C. termitidis on cellobiose, xylose, xylan, and α-cellulose.
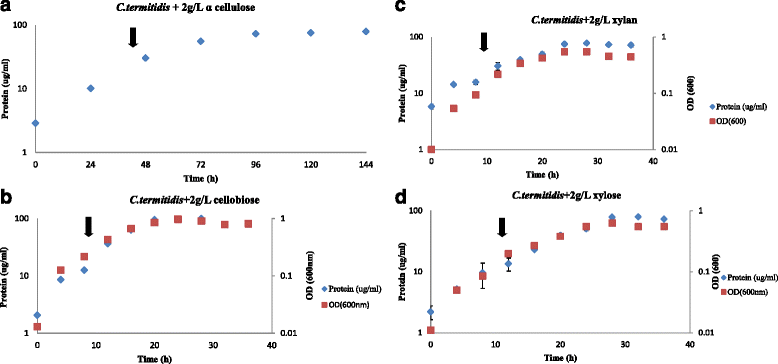
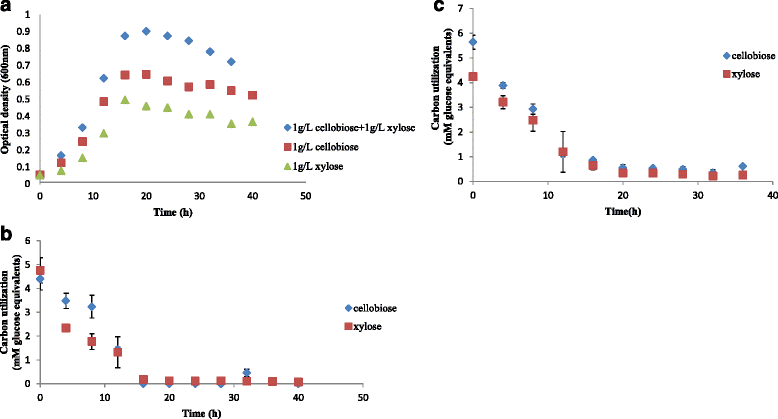
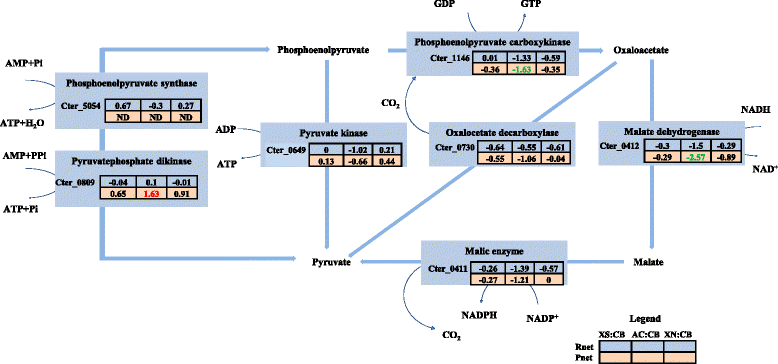
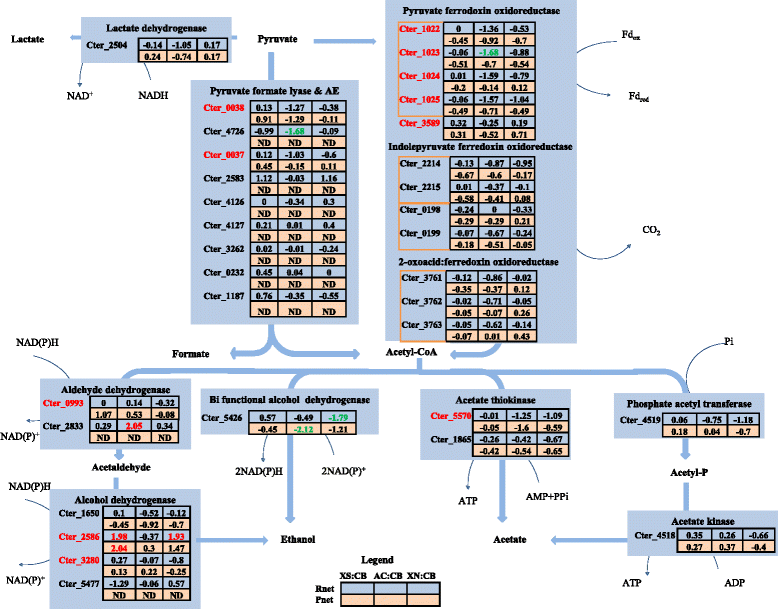
Results: Correlation of transcriptome and proteome data with growth and fermentation profiles identified putative carbon-catabolism pathways in C. termitidis. The majority of the proteins associated with central metabolism were detected in high abundance. While major differences were not observed in gene and gene-product expression for enzymes associated with metabolic pathways under the different substrate conditions, xylulokinase and xylose isomerase of the pentose phosphate pathway were found to be highly up-regulated on five carbon sugars compared to hexoses. In addition, genes and gene-products associated with a variety of cellulosome and non-cellulosome associated CAZymes were found to be differentially expressed. Specifically, genes for cellulosomal enzymes and components were highly expressed on α-cellulose, while xylanases and glucosidases were up-regulated on 5 carbon sugars with respect to cellobiose. Chitinase and cellobiophosphorylases were the predominant CAZymes expressed on cellobiose. In addition to growth on xylan, the simultaneous consumption of two important lignocellulose constituents, cellobiose and xylose was also demonstrated.
Conclusion: There are little changes in core-metabolic pathways under the different carbon sources compared. The most significant differences were found to be associated with the CAZymes, as well as specific up regulation of some key components of the pentose phosphate pathway in the presence of xylose and xylan. This study has enhanced our understanding of the physiology and metabolism of C. termitidis, and provides a foundation for future studies on metabolic engineering to optimize biofuel production from natural biomass.
Keywords: Biofuel; CAZymes; Clostridium termitidis; Metabolism; Quantitative proteomics; RNAseq.
Figures


Similar articles
-
Quantitative proteomic analysis of the cellulolytic system of Clostridium termitidis CT1112 reveals distinct protein expression profiles upon growth on α-cellulose and cellobiose.J Proteomics. 2015 Jul 1;125:41-53. doi: 10.1016/j.jprot.2015.04.026. Epub 2015 May 7. J Proteomics. 2015. PMID: 25957533
-
Comparative analysis of carbohydrate active enzymes in Clostridium termitidis CT1112 reveals complex carbohydrate degradation ability.PLoS One. 2014 Aug 7;9(8):e104260. doi: 10.1371/journal.pone.0104260. eCollection 2014. PLoS One. 2014. PMID: 25101643 Free PMC article.
-
Comparative Genomics of Core Metabolism Genes of Cellulolytic and Non-cellulolytic Clostridium Species.Adv Biochem Eng Biotechnol. 2016;156:79-112. doi: 10.1007/10_2015_5007. Adv Biochem Eng Biotechnol. 2016. PMID: 26907553
-
Cellulolytic and hemicellulolytic capacity of Acetivibrio clariflavus.Appl Microbiol Biotechnol. 2025 Apr 28;109(1):105. doi: 10.1007/s00253-025-13471-9. Appl Microbiol Biotechnol. 2025. PMID: 40295343 Free PMC article. Review.
-
Simultaneous bioconversion of cellulose and hemicellulose to ethanol.Crit Rev Biotechnol. 1998;18(4):295-331. doi: 10.1080/0738-859891224185. Crit Rev Biotechnol. 1998. PMID: 9887507 Review.
Cited by
-
Whole Proteome Analyses on Ruminiclostridium cellulolyticum Show a Modulation of the Cellulolysis Machinery in Response to Cellulosic Materials with Subtle Differences in Chemical and Structural Properties.PLoS One. 2017 Jan 23;12(1):e0170524. doi: 10.1371/journal.pone.0170524. eCollection 2017. PLoS One. 2017. PMID: 28114419 Free PMC article.
-
Unraveling essential cellulosomal components of the (Pseudo)Bacteroides cellulosolvens reveals an extensive reservoir of novel catalytic enzymes.Biotechnol Biofuels. 2019 May 9;12:115. doi: 10.1186/s13068-019-1447-2. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31086567 Free PMC article.
-
Bioconversion of Lignocellulosic Biomass into Value Added Products under Anaerobic Conditions: Insight into Proteomic Studies.Int J Mol Sci. 2021 Nov 12;22(22):12249. doi: 10.3390/ijms222212249. Int J Mol Sci. 2021. PMID: 34830131 Free PMC article. Review.
-
Developing Clostridia as Cell Factories for Short- and Medium-Chain Ester Production.Front Bioeng Biotechnol. 2021 Jun 7;9:661694. doi: 10.3389/fbioe.2021.661694. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34164382 Free PMC article. Review.
-
Combining transcriptomic and metabolomic insights into carbohydrate utilization by Ruminiclostridium papyrosolvens DSM2782.Biotechnol Biofuels Bioprod. 2025 Feb 22;18(1):22. doi: 10.1186/s13068-025-02619-4. Biotechnol Biofuels Bioprod. 2025. PMID: 39987219 Free PMC article.
References
-
- Wei H, Fu Y, Magnusson L, Baker JO, Maness P-C, Xu Q, Yang S, Bowersox A, Bogorad I, Wang W. Comparison of transcriptional profiles of Clostridium thermocellum grown on cellobiose and pretreated yellow poplar using RNA-Seq. Front Microbiol. 2014;5:1–16. doi: 10.3389/fmicb.2014.00142. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

