The Structure-Specific Recognition Protein 1 Associates with Lens Epithelium-Derived Growth Factor Proteins and Modulates HIV-1 Replication
- PMID: 27216501
- PMCID: PMC4938748
- DOI: 10.1016/j.jmb.2016.05.013
The Structure-Specific Recognition Protein 1 Associates with Lens Epithelium-Derived Growth Factor Proteins and Modulates HIV-1 Replication
Abstract
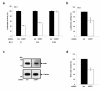
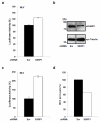
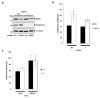
The lens epithelium-derived growth factor p75 (LEDGF/p75) is a chromatin-bound protein essential for efficient lentiviral integration. Genome-wide studies have located LEDGF/p75 inside actively transcribed genes where it mediates lentiviral integration. Although its role in HIV-1 integration is clearly established, the role of LEDGF/p75-associated proteins in HIV-1 infection remains unexplored. Using protein-protein interaction assays, we demonstrated that LEDGF/p75 complexes with a chromatin-remodeling complex facilitates chromatin transcription (FACT), a heterodimer of the structure-specific recognition protein 1 (SSRP1) and the human homolog of suppressor of Ty 16 (hSpt16). Detailed analysis of the interaction of LEDGF/p75 with the FACT complex indicates that LEDGF/p75 interacts with SSRP1 in an hSpt16-independent manner that requires the PWWP domain of LEDGF proteins and the HMG domain of SSRP1. Functional characterizations demonstrate a LEDGF/p75-independent role of SSRP1 in the regulation of HIV-1 replication. shRNA-mediated partial knockdown of SSRP1 reduces HIV-1 infection, but not Murine Leukemia Virus, in human CD4(+) T cells. Similarly, SSRP1 knockdown affects infection by HIV-1-derived viruses that express genes from the viral LTR but not from an internal immediate-early CMV promoter, suggesting a role of SSRP1 in LTR-driven gene expression but not in viral DNA integration. Together, our data demonstrate for the first time the association of LEDGF proteins with the FACT complex and give further support to a role of SSRP1 in HIV-1 infection.
Keywords: FACT complex; HIV-1 cofactor; HIV-1 replication; HMG domain; PWWP domain.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures


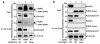

Similar articles
-
Role of the PWWP domain of lens epithelium-derived growth factor (LEDGF)/p75 cofactor in lentiviral integration targeting.J Biol Chem. 2011 Dec 2;286(48):41812-41826. doi: 10.1074/jbc.M111.255711. Epub 2011 Oct 10. J Biol Chem. 2011. Retraction in: J Biol Chem. 2018 Jan 5;293(1):114. doi: 10.1074/jbc.W117.001458. PMID: 21987578 Free PMC article. Retracted.
-
Characterization of the HIV-1 integrase chromatin- and LEDGF/p75-binding abilities by mutagenic analysis within the catalytic core domain of integrase.Virol J. 2010 Mar 23;7:68. doi: 10.1186/1743-422X-7-68. Virol J. 2010. PMID: 20331877 Free PMC article.
-
HIV-1 integrase modulates the interaction of the HIV-1 cellular cofactor LEDGF/p75 with chromatin.Retrovirology. 2011 Apr 21;8:27. doi: 10.1186/1742-4690-8-27. Retrovirology. 2011. PMID: 21510906 Free PMC article.
-
The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication.PLoS Pathog. 2008 Mar 28;4(3):e1000046. doi: 10.1371/journal.ppat.1000046. PLoS Pathog. 2008. PMID: 18369482 Free PMC article. Review.
-
Protein-protein and protein-chromatin interactions of LEDGF/p75 as novel drug targets.Drug Discov Today Technol. 2017 Jun;24:25-31. doi: 10.1016/j.ddtec.2017.11.002. Epub 2017 Nov 23. Drug Discov Today Technol. 2017. PMID: 29233296 Review.
Cited by
-
Current Strategies for Elimination of HIV-1 Latent Reservoirs Using Chemical Compounds Targeting Host and Viral Factors.AIDS Res Hum Retroviruses. 2019 Jan;35(1):1-24. doi: 10.1089/AID.2018.0153. Epub 2018 Dec 12. AIDS Res Hum Retroviruses. 2019. PMID: 30351168 Free PMC article. Review.
-
The Emerging Roles of the Stress Epigenetic Reader LEDGF/p75 in Cancer Biology and Therapy Resistance: Mechanisms and Targeting Opportunities.Cancers (Basel). 2024 Nov 26;16(23):3957. doi: 10.3390/cancers16233957. Cancers (Basel). 2024. PMID: 39682146 Free PMC article. Review.
-
Curaxin CBL0100 Blocks HIV-1 Replication and Reactivation through Inhibition of Viral Transcriptional Elongation.Front Microbiol. 2017 Oct 17;8:2007. doi: 10.3389/fmicb.2017.02007. eCollection 2017. Front Microbiol. 2017. PMID: 29089933 Free PMC article.
-
Retroviral integrase: Structure, mechanism, and inhibition.Enzymes. 2021;50:249-300. doi: 10.1016/bs.enz.2021.06.007. Epub 2021 Aug 23. Enzymes. 2021. PMID: 34861940 Free PMC article.
-
Modulation of chromatin structure by the FACT histone chaperone complex regulates HIV-1 integration.Retrovirology. 2017 Jul 28;14(1):39. doi: 10.1186/s12977-017-0363-4. Retrovirology. 2017. PMID: 28754126 Free PMC article.
References
-
- Ge H, Si Y, Wolffe AP. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell. 1998;2:751–9. - PubMed
-
- Kubo E, Singh DP, Fatma N, Shinohara T, Zelenka P, Reddy VN, Chylack LT. Cellular distribution of lens epithelium-derived growth factor (LEDGF) in the rat eye: loss of LEDGF from nuclei of differentiating cells. Histochem Cell Biol. 2003;119:289–99. - PubMed
-
- Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol. 2006;360:760–73. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

