Elevated variant density around SV breakpoints in germline lineage lends support to error-prone replication hypothesis
- PMID: 27216746
- PMCID: PMC4937565
- DOI: 10.1101/gr.205484.116
Elevated variant density around SV breakpoints in germline lineage lends support to error-prone replication hypothesis
Abstract
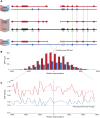
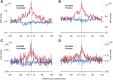
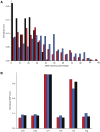
Copy number variants (CNVs) are a class of structural variants that may involve complex genomic rearrangements (CGRs) and are hypothesized to have additional mutations around their breakpoints. Understanding the mechanisms underlying CNV formation is fundamental for understanding the repair and mutation mechanisms in cells, thereby shedding light on evolution, genomic disorders, cancer, and complex human traits. In this study, we used data from the 1000 Genomes Project to analyze hundreds of loci harboring heterozygous germline deletions in the subjects NA12878 and NA19240. By utilizing synthetic long-read data (longer than 2 kbp) in combination with high coverage short-read data and, in parallel, by comparing with parental genomes, we interrogated the phasing of these deletions with the flanking tens of thousands of heterozygous SNPs and indels. We found that the density of SNPs/indels flanking the breakpoints of deletions (in-phase variants) is approximately twice as high as the corresponding density for the variants on the haplotype without deletion (out-of-phase variants). This fold change was even larger for the subset of deletions with signatures of replication-based mechanism of formation. The allele frequency (AF) spectrum for deletions is enriched for rare events; and the AF spectrum for in-phase SNPs is shifted toward this deletion spectrum, thus offering evidence consistent with the concomitance of the in-phase SNPs/indels with the deletion events. These findings therefore lend support to the hypothesis that the mutational mechanisms underlying CNV formation are error prone. Our results could also be relevant for resolving mutation-rate discrepancies in human and to explain kataegis.
© 2016 Dhokarh and Abyzov; Published by Cold Spring Harbor Laboratory Press.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
