CRISPR/Cas9 for Human Genome Engineering and Disease Research
- PMID: 27216776
- PMCID: PMC10216851
- DOI: 10.1146/annurev-genom-083115-022258
CRISPR/Cas9 for Human Genome Engineering and Disease Research
Abstract
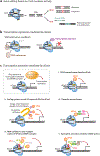
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) system, a versatile RNA-guided DNA targeting platform, has been revolutionizing our ability to modify, manipulate, and visualize the human genome, which greatly advances both biological research and therapeutics development. Here, we review the current development of CRISPR/Cas9 technologies for gene editing, transcription regulation, genome imaging, and epigenetic modification. We discuss the broad application of this system to the study of functional genomics, especially genome-wide genetic screening, and to therapeutics development, including establishing disease models, correcting defective genetic mutations, and treating diseases.
Keywords: gene editing; gene regulation; gene therapy; genetic screening; human diseases.
Figures


References
-
- Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, et al. 2015. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 10:1422–32 - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–12 - PubMed
-
- Beerli RR, Barbas CF III. 2002. Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol 20:135–41 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

