Tet-On Systems For Doxycycline-inducible Gene Expression
- PMID: 27216914
- PMCID: PMC5070417
- DOI: 10.2174/1566523216666160524144041
Tet-On Systems For Doxycycline-inducible Gene Expression
Abstract
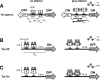
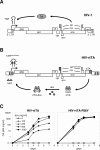
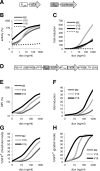
The tetracycline-controlled Tet-Off and Tet-On gene expression systems are used to regulate the activity of genes in eukaryotic cells in diverse settings, varying from basic biological research to biotechnology and gene therapy applications. These systems are based on regulatory elements that control the activity of the tetracycline-resistance operon in bacteria. The Tet-Off system allows silencing of gene expression by administration of tetracycline (Tc) or tetracycline-derivatives like doxycycline (dox), whereas the Tet-On system allows activation of gene expression by dox. Since the initial design and construction of the original Tet-system, these bacterium-derived systems have been significantly improved for their function in eukaryotic cells. We here review how a dox-controlled HIV-1 variant was designed and used to greatly improve the activity and dox-sensitivity of the rtTA transcriptional activator component of the Tet-On system. These optimized rtTA variants require less dox for activation, which will reduce side effects and allow gene control in tissues where a relatively low dox level can be reached, such as the brain.
Figures



References
-
- Gossen M., Bujard H. Tetracyclines in the control of gene expression in eukaryotes. In: Nelson M., Hillen W., Greenwald R.A., editors. Tetracyclines in biology, chemistry and medicine. Basel: Birkhäuser Verlag; 2001. pp. 139–157. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

