HtrA Is Important for Stress Resistance and Virulence in Haemophilus parasuis
- PMID: 27217419
- PMCID: PMC4962635
- DOI: 10.1128/IAI.00147-16
HtrA Is Important for Stress Resistance and Virulence in Haemophilus parasuis
Abstract
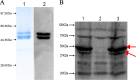
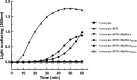
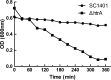
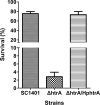
Haemophilus parasuis is an opportunistic pathogen that causes Glässer's disease in swine, with polyserositis, meningitis, and arthritis. The high-temperature requirement A (HtrA)-like protease, which is involved in protein quality control, has been reported to be a virulence factor in many pathogens. In this study, we showed that HtrA of H. parasuis (HpHtrA) exhibited both chaperone and protease activities. Finally, nickel import ATP-binding protein (NikE), periplasmic dipeptide transport protein (DppA), and outer membrane protein A (OmpA) were identified as proteolytic substrates for HpHtrA. The protease activity reached its maximum at 40°C in a time-dependent manner. Disruption of the htrA gene from strain SC1401 affected tolerance to temperature stress and resistance to complement-mediated killing. Furthermore, increased autoagglutination and biofilm formation were detected in the htrA mutant. In addition, the htrA mutant was significantly attenuated in virulence in the murine model of infection. Together, these data demonstrate that HpHtrA plays an important role in the virulence of H. parasuis.
Copyright © 2016 Zhang et al.
Figures





Similar articles
-
Deletion of the vacJ gene affects the biology and virulence in Haemophilus parasuis serovar 5.Gene. 2017 Mar 1;603:42-53. doi: 10.1016/j.gene.2016.12.009. Epub 2016 Dec 14. Gene. 2017. PMID: 27988234
-
The AI-2/luxS Quorum Sensing System Affects the Growth Characteristics, Biofilm Formation, and Virulence of Haemophilus parasuis.Front Cell Infect Microbiol. 2019 Mar 19;9:62. doi: 10.3389/fcimb.2019.00062. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 30941317 Free PMC article.
-
HtrA is involved in stress response and adhesion in Glaesserella parasuis serovar 5 strain Nagasaki.Vet Microbiol. 2023 Jul;282:109748. doi: 10.1016/j.vetmic.2023.109748. Epub 2023 Apr 18. Vet Microbiol. 2023. PMID: 37120968
-
Advances in the quest for virulence factors of Haemophilus parasuis.Vet J. 2013 Dec;198(3):571-6. doi: 10.1016/j.tvjl.2013.08.027. Epub 2013 Sep 4. Vet J. 2013. PMID: 24084037 Review.
-
Update on the pathogenesis of Haemophilus parasuis infection and virulence factors.Vet Microbiol. 2014 Jan 10;168(1):1-7. doi: 10.1016/j.vetmic.2013.07.027. Epub 2013 Aug 12. Vet Microbiol. 2014. PMID: 23972951 Review.
Cited by
-
Arginine Decarboxylase Is Essential for Pneumococcal Stress Responses.Pathogens. 2021 Mar 2;10(3):286. doi: 10.3390/pathogens10030286. Pathogens. 2021. PMID: 33801541 Free PMC article.
-
A TolC-Like Protein of Actinobacillus pleuropneumoniae Is Involved in Antibiotic Resistance and Biofilm Formation.Front Microbiol. 2016 Oct 24;7:1618. doi: 10.3389/fmicb.2016.01618. eCollection 2016. Front Microbiol. 2016. PMID: 27822201 Free PMC article.
-
Modeling Co-Infection by Streptococcus suis and Haemophilus parasuis Reveals Influences on Biofilm Formation and Host Response.Animals (Basel). 2023 Apr 29;13(9):1511. doi: 10.3390/ani13091511. Animals (Basel). 2023. PMID: 37174548 Free PMC article.
-
Baicalin Inhibits Haemophilus Parasuis-Induced High-Mobility Group Box 1 Release during Inflammation.Int J Mol Sci. 2018 Apr 27;19(5):1307. doi: 10.3390/ijms19051307. Int J Mol Sci. 2018. PMID: 29702580 Free PMC article.
-
A streptomycin resistance marker in H. parasuis based on site-directed mutations in rpsL gene to perform unmarked in-frame mutations and to verify natural transformation.PeerJ. 2018 Jan 11;6:e4253. doi: 10.7717/peerj.4253. eCollection 2018. PeerJ. 2018. PMID: 29340249 Free PMC article.
References
-
- Zhang L, Wen Y, Li Y, Wei X, Yan X, Wen X, Wu R, Huang X, Huang Y, Yan Q, Liu M, Cao S. 2014. Comparative proteomic analysis of the membrane proteins of two Haemophilus parasuis strains to identify proteins that may help in habitat adaptation and pathogenesis. Proteome Sci 12:38. doi:10.1186/1477-5956-12-38. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

