A Laser Dissection-RNAseq Analysis Highlights the Activation of Cytokinin Pathways by Nod Factors in the Medicago truncatula Root Epidermis
- PMID: 27217496
- PMCID: PMC4936592
- DOI: 10.1104/pp.16.00711
A Laser Dissection-RNAseq Analysis Highlights the Activation of Cytokinin Pathways by Nod Factors in the Medicago truncatula Root Epidermis
Abstract
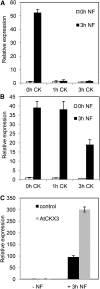
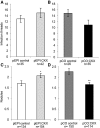
Nod factors (NFs) are lipochitooligosaccharidic signal molecules produced by rhizobia, which play a key role in the rhizobium-legume symbiotic interaction. In this study, we analyzed the gene expression reprogramming induced by purified NF (4 and 24 h of treatment) in the root epidermis of the model legume Medicago truncatula Tissue-specific transcriptome analysis was achieved by laser-capture microdissection coupled to high-depth RNA sequencing. The expression of 17,191 genes was detected in the epidermis, among which 1,070 were found to be regulated by NF addition, including previously characterized NF-induced marker genes. Many genes exhibited strong levels of transcriptional activation, sometimes only transiently at 4 h, indicating highly dynamic regulation. Expression reprogramming affected a variety of cellular processes, including perception, signaling, regulation of gene expression, as well as cell wall, cytoskeleton, transport, metabolism, and defense, with numerous NF-induced genes never identified before. Strikingly, early epidermal activation of cytokinin (CK) pathways was indicated, based on the induction of CK metabolic and signaling genes, including the CRE1 receptor essential to promote nodulation. These transcriptional activations were independently validated using promoter:β-glucuronidase fusions with the MtCRE1 CK receptor gene and a CK response reporter (TWO COMPONENT SIGNALING SENSOR NEW). A CK pretreatment reduced the NF induction of the EARLY NODULIN11 (ENOD11) symbiotic marker, while a CK-degrading enzyme (CYTOKININ OXIDASE/DEHYDROGENASE3) ectopically expressed in the root epidermis led to increased NF induction of ENOD11 and nodulation. Therefore, CK may play both positive and negative roles in M. truncatula nodulation.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Andrio E, Marino D, Marmeys A, de Segonzac MD, Damiani I, Genre A, Huguet S, Frendo P, Puppo A, Pauly N (2013) Hydrogen peroxide-regulated genes in the Medicago truncatula-Sinorhizobium meliloti symbiosis. New Phytol 198: 179–189 - PubMed
-
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 - PubMed
-
- Antolín-Llovera M, Ried MK, Parniske M (2014) Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr Biol 24: 422–427 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

