Interferon-driven alterations of the host's amino acid metabolism in the pathogenesis of typhoid fever
- PMID: 27217537
- PMCID: PMC4886356
- DOI: 10.1084/jem.20151025
Interferon-driven alterations of the host's amino acid metabolism in the pathogenesis of typhoid fever
Abstract
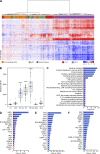
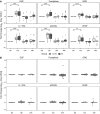
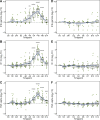
Enteric fever, caused by Salmonella enterica serovar Typhi, is an important public health problem in resource-limited settings and, despite decades of research, human responses to the infection are poorly understood. In 41 healthy adults experimentally infected with wild-type S. Typhi, we detected significant cytokine responses within 12 h of bacterial ingestion. These early responses did not correlate with subsequent clinical disease outcomes and likely indicate initial host-pathogen interactions in the gut mucosa. In participants developing enteric fever after oral infection, marked transcriptional and cytokine responses during acute disease reflected dominant type I/II interferon signatures, which were significantly associated with bacteremia. Using a murine and macrophage infection model, we validated the pivotal role of this response in the expression of proteins of the host tryptophan metabolism during Salmonella infection. Corresponding alterations in tryptophan catabolites with immunomodulatory properties in serum of participants with typhoid fever confirmed the activity of this pathway, and implicate a central role of host tryptophan metabolism in the pathogenesis of typhoid fever.
© 2016 Blohmke et al.
Figures




References
-
- Banchereau R., Jordan-Villegas A., Ardura M., Mejias A., Baldwin N., Xu H., Saye E., Rossello-Urgell J., Nguyen P., Blankenship D., et al. . 2012. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One. 7:e34390 10.1371/journal.pone.0034390 - DOI - PMC - PubMed
-
- Blumenthal A., Nagalingam G., Huch J.H., Walker L., Guillemin G.J., Smythe G.A., Ehrt S., Britton W.J., and Saunders B.M.M.. 2012. M. tuberculosis induces potent activation of IDO-1, but this is not essential for the immunological control of infection. PLoS One. 7:e37314 10.1371/journal.pone.0037314 - DOI - PMC - PubMed
-
- Bucasas K.L., Franco L.M., Shaw C.A., Bray M.S., Wells J.M., Niño D., Arden N., Quarles J.M., Couch R.B., and Belmont J.W.. 2011. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J. Infect. Dis. 203:921–929. 10.1093/infdis/jiq156 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

