Association of primary open-angle glaucoma with mitochondrial variants and haplogroups common in African Americans
- PMID: 27217714
- PMCID: PMC4872278
Association of primary open-angle glaucoma with mitochondrial variants and haplogroups common in African Americans
Abstract
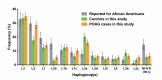
Purpose: To estimate the population frequencies of all common mitochondrial variants and ancestral haplogroups among 1,999 subjects recruited for the Primary Open-Angle African American Glaucoma Genetics (POAAGG) Study, including 1,217 primary open-angle glaucoma (POAG) cases and 782 controls, and to identify ancestral subpopulations and mitochondrial mutations as potential risk factors for POAG susceptibility.
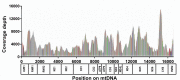
Methods: Subject classification by characteristic glaucomatous optic nerve findings and corresponding visual field defects, as defined by enrolling glaucoma specialists, stereo disc photography, phlebotomy, extraction of total DNA from peripheral blood or saliva, DNA quantification and normalization, PCR amplification of whole mitochondrial genomes, Ion Torrent deep semiconductor DNA sequencing on DNA pools ("Pool-seq"), Sanger sequencing of 3,479 individual mitochondrial DNAs, and bioinformatic analysis.
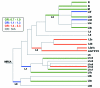
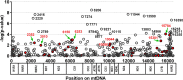
Results: The distribution of common African haplogroups within the POAAGG study population was broadly similar to prior surveys of African Americans. However, the POAG case population was found to be enriched in L1c2 haplogroups, which are defined in part by missense mutations m.6150G>A (Val83Ile, odds ratio [OR] 1.8, p=0.01), m.6253C>T (Met117Thr, rs200165736, OR 1.6, p=0.04), and m.6480G>A (Val193Ile, rs199476128, OR 4.6, p=0.04) in the cytochrome c oxidase subunit 1 (MT-CO1) gene and by a variant, m.2220A>G (OR 2.0, p=0.01), in MT-RNR2, which encodes the mitochondrial ribosomal 16s RNA gene. L2 haplogroups were predicted to be overrepresented in the POAG case population by Pool-seq, and the difference was confirmed to be significant with Sanger sequencing, that targeted the L2-associated variants m.2416T>C (rs28358580, OR 1.2, p=0.02) and m.2332C>T (OR 1.2, p=.02) in MT-RNR2. Another variant within MT-RNR2, m.3010G>A (rs3928306), previously implicated in sensitivity to the optic neuropathy-associated antibiotic linezolid, and arising on D4 and J1 lineages, associated with Leber hereditary optic neuropathy (LHON) severity, was confirmed to be common (>5%) but was not significantly enriched in the POAG cases. Two variants linked to the composition of the gut microbiome, m.15784T>C (rs527236194, haplogroup L2a1) and m.16390G>A (rs41378955, L2 haplogroups), were also enriched in the case DNA pools.
Conclusions: These results implicate African mtDNA haplogroups L1c2, L1c2b, and L2 as risk factors for POAG. Approximately one in four African Americans have these mitochondrial ancestries, which may contribute to their elevated glaucoma risk. These haplogroups are defined in part by ancestral variants in the MT-RNR2 and/or MT-CO1 genes, several of which have prior disease associations, such as MT-CO1 missense variants that have been implicated in prostate cancer.
Figures

References
-
- Tielsch JM, Katz J, Singh K, Quigley HA, Gottsch JD, Javitt J, Sommer A. A population-based evaluation of glaucoma screening: The baltimore eye survey. Am J Epidemiol. 1991;134:1102–10. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&lis... - PubMed
-
- Osborne NN, Alvarez CN, del Olmo Aguado S. Targeting mitochondrial dysfunction as in aging and glaucoma. Drug Discov Today. 2014;19:1613–22. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&lis... - PubMed
-
- Gomez-Duran A, Pacheu-Grau D, Martinez-Romero I, Lopez-Gallardo E, Lopez-Perez MJ, Montoya J, Ruiz-Pesini E. Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of leber’s hereditary optic neuropathy. Biochim Biophys Acta. 2012;2012:1216–22. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&lis... - PubMed
-
- Ghelli A, Porcelli AM, Zanna C, Vidoni S, Mattioli S, Barbieri A, Iommarini L, Pala M, Achilli A, Torroni A, Rugolo M, Carelli V. The background of mitochondrial DNA haplogroup J increases the sensitivity of leber’s hereditary optic neuropathy cells to 2,5-hexanedione toxicity. PLoS One. 2009;4:e7922. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&lis... - PMC - PubMed
-
- Lascaratos G, Garway-Heath DF, Willoughby CE, Chau KY, Schapira AH. Mitochondrial dysfunction in glaucoma: Understanding genetic influences. Mitochondrion. 2012;12:202–12. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&lis... - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

