A comparative reference study for the validation of HLA-matching algorithms in the search for allogeneic hematopoietic stem cell donors and cord blood units
- PMID: 27219013
- PMCID: PMC5089599
- DOI: 10.1111/tan.12817
A comparative reference study for the validation of HLA-matching algorithms in the search for allogeneic hematopoietic stem cell donors and cord blood units
Abstract
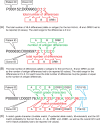
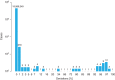
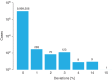
The accuracy of human leukocyte antigen (HLA)-matching algorithms is a prerequisite for the correct and efficient identification of optimal unrelated donors for patients requiring hematopoietic stem cell transplantation. The goal of this World Marrow Donor Association study was to validate established matching algorithms from different international donor registries by challenging them with simulated input data and subsequently comparing the output. This experiment addressed three specific aspects of HLA matching using different data sets for tasks of increasing complexity. The first two tasks targeted the traditional matching approach identifying discrepancies between patient and donor HLA genotypes by counting antigen and allele differences. Contemporary matching procedures predicting the probability for HLA identity using haplotype frequencies were addressed by the third task. In each task, the identified disparities between the results of the participating computer programs were analyzed, classified and quantified. This study led to a deep understanding of the algorithms participating and finally produced virtually identical results. The unresolved discrepancies total to less than 1%, 4% and 2% for the three tasks and are mostly because of individual decisions in the design of the programs. Based on these findings, reference results for the three input data sets were compiled that can be used to validate future matching algorithms and thus improve the quality of the global donor search process.
Keywords: donor registry; hematopoietic stem cell transplantation; human leukocyte antigen; immunogenetics; matching algorithm; patient-donor matching.
© 2016 The Authors. HLA published by John Wiley & Sons Ltd.
Figures
Similar articles
-
Identification of a 10/10 matched donor for patients with an uncommon haplotype is unlikely.HLA. 2017 Feb;89(2):77-81. doi: 10.1111/tan.12946. HLA. 2017. PMID: 28102042
-
Comparative validation of computer programs for haplotype frequency estimation from donor registry data.Tissue Antigens. 2013 Aug;82(2):93-105. doi: 10.1111/tan.12160. Tissue Antigens. 2013. PMID: 23849067
-
Availability of HLA-allele-matched unrelated donors and registry size: Estimation from haplotype frequency in the Italian population.Hum Immunol. 2021 Oct;82(10):758-766. doi: 10.1016/j.humimm.2021.07.012. Epub 2021 Aug 2. Hum Immunol. 2021. PMID: 34353675
-
Maximizing optimal hematopoietic stem cell donor selection from registries of unrelated adult volunteers.Tissue Antigens. 2003 Jun;61(6):415-24. doi: 10.1034/j.1399-0039.2003.00096.x. Tissue Antigens. 2003. PMID: 12823765 Review.
-
In celebration of Ruggero Ceppellini: HLA in transplantation.HLA. 2017 Feb;89(2):71-76. doi: 10.1111/tan.12955. HLA. 2017. PMID: 28102037 Free PMC article. Review.
Cited by
-
Estimation of German KIR Allele Group Haplotype Frequencies.Front Immunol. 2020 Mar 12;11:429. doi: 10.3389/fimmu.2020.00429. eCollection 2020. Front Immunol. 2020. PMID: 32226430 Free PMC article.
-
Directionality of non-permissive HLA-DPB1 T-cell epitope group mismatches does not improve clinical risk stratification in 8/8 matched unrelated donor hematopoietic cell transplantation.Bone Marrow Transplant. 2017 Sep;52(9):1280-1287. doi: 10.1038/bmt.2017.96. Epub 2017 Jun 5. Bone Marrow Transplant. 2017. PMID: 28581467 Free PMC article.
-
Hap-E Search 2.0: Improving the Performance of a Probabilistic Donor-Recipient Matching Algorithm Based on Haplotype Frequencies.Front Med (Lausanne). 2020 Feb 18;7:32. doi: 10.3389/fmed.2020.00032. eCollection 2020. Front Med (Lausanne). 2020. PMID: 32133364 Free PMC article.
-
Hapl-o-Mat: open-source software for HLA haplotype frequency estimation from ambiguous and heterogeneous data.BMC Bioinformatics. 2017 May 30;18(1):284. doi: 10.1186/s12859-017-1692-y. BMC Bioinformatics. 2017. PMID: 28558647 Free PMC article.
-
Common and well-documented HLA alleles of German stem cell donors by haplotype frequency estimation.HLA. 2018 Oct;92(4):206-214. doi: 10.1111/tan.13378. HLA. 2018. PMID: 30117303 Free PMC article.
References
-
- Fürst D, Müller CR, Vucinic V et al. High‐resolution HLA matching in hematopoietic stem cell transplantation: a retrospective collaborative analysis. Blood 2013: 122: 3220–9. - PubMed
-
- Lee SJ, Klein J, Haagenson M et al. High‐resolution donor‐recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood 2007: 110: 4576–83. - PubMed
-
- Hurley CK, Maiers M, Marsh SGE, Oudshoorn M. Overview of registries, HLA typing and diversity, and search algorithms. Tissue Antigens 2007: 69 (Suppl 1): 3–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

