Fishing for Function in the Human Gene Pool
- PMID: 27220646
- PMCID: PMC4912909
- DOI: 10.1016/j.tig.2016.05.002
Fishing for Function in the Human Gene Pool
Abstract
Identification and characterization of causal non-coding variants in human genomes is challenging and requires substantial experimental resources. A new study by Tehranchi et al. describes a cost-effective approach for accurate mapping of molecular quantitative trait loci (QTLs) from pooled samples, a powerful way to link disease-associated changes to molecular functions.
Keywords: GWAS; QTL; cis-regulation; non-coding DNA.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interests.
Figures
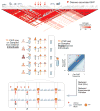
Comment on
-
Pooled ChIP-Seq Links Variation in Transcription Factor Binding to Complex Disease Risk.Cell. 2016 Apr 21;165(3):730-41. doi: 10.1016/j.cell.2016.03.041. Epub 2016 Apr 14. Cell. 2016. PMID: 27087447 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

