A Buffered Alcohol-Based Fixative for Histomorphologic and Molecular Applications
- PMID: 27221702
- PMCID: PMC4931761
- DOI: 10.1369/0022155416649579
A Buffered Alcohol-Based Fixative for Histomorphologic and Molecular Applications
Abstract
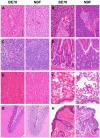
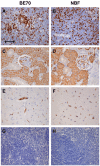
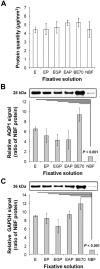
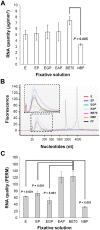
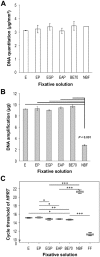
Formalin-fixed paraffin-embedded (FFPE) tissue is the predominant preparation for diagnostic histopathological evaluation and increasingly the biospecimen on which molecular diagnostics are performed. However, formalin is carcinogenic and results in cross-linking of proteins and nicking and alterations of nucleic acids. Alternative fixatives, including 70% ethanol, improved biomolecular integrity; however, they have yet to replace neutral-buffered formalin (NBF). Herein, we describe the phosphate-buffered ethanol 70% (BE70) fixative. The histomorphology of BE70-fixed tissue is very similar to that of NBF; however, it is a non-cross-linking fixative and lacks the carcinogenic profile of formaldehyde-based fixatives. RNA isolated from tissue fixed in BE70 was of substantially higher quality and quantity than that was recovered from formalin-fixed tissue. Furthermore, the BE70 fixative showed excellent RNA and DNA integrity compared with that of NBF fixative based on real-time polymerase chain reaction analysis results. Immunohistochemical staining was similar for the antigen tested. In conclusion, BE70 is a non-cross-linking fixative that is superior to NBF and 70% ethanol with reference to biomolecule recovery and quality from paraffin-embedded tissue. Additional studies to compare the histomorphologic and immunohistochemical performance and utility in a clinical setting are required.
Keywords: RNA integrity; alcohol; fixation; formalin; histomorphology; paraffin embedded; real-time RT-PCR; tissue.
© 2016 The Histochemical Society.
Conflict of interest statement
Figures


Comment in
-
Formulation and pH of the Buffered Ethanol Fixative BE70.J Histochem Cytochem. 2017 Apr;65(4):251-252. doi: 10.1369/0022155416687279. J Histochem Cytochem. 2017. PMID: 28347266 Free PMC article. No abstract available.
-
Buffered Ethanol Fixative.J Histochem Cytochem. 2017 Apr;65(4):251. doi: 10.1369/0022155416687278. J Histochem Cytochem. 2017. PMID: 28347267 Free PMC article. No abstract available.
Similar articles
-
The Application of Guanidinium to Improve Biomolecule Quality in Fixed, Paraffin-embedded Tissue.J Histochem Cytochem. 2023 Feb;71(2):87-101. doi: 10.1369/00221554231159451. Epub 2023 Mar 4. J Histochem Cytochem. 2023. PMID: 36869703 Free PMC article.
-
Histomorphological and Molecular Assessments of the Fixation Times Comparing Formalin and Ethanol-Based Fixatives.J Histochem Cytochem. 2018 Feb;66(2):121-135. doi: 10.1369/0022155417741467. Epub 2017 Nov 10. J Histochem Cytochem. 2018. PMID: 29125916 Free PMC article.
-
Formulation and pH of the Buffered Ethanol Fixative BE70.J Histochem Cytochem. 2017 Apr;65(4):251-252. doi: 10.1369/0022155416687279. J Histochem Cytochem. 2017. PMID: 28347266 Free PMC article. No abstract available.
-
Effects of fixative and fixation time on the extraction and polymerase chain reaction amplification of RNA from paraffin-embedded tissue. Comparison of two housekeeping gene mRNA controls.Diagn Mol Pathol. 1994 Sep;3(3):148-55. doi: 10.1097/00019606-199409000-00003. Diagn Mol Pathol. 1994. PMID: 7981889 Review.
-
A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen?Arch Pathol Lab Med. 2014 Nov;138(11):1520-30. doi: 10.5858/arpa.2013-0691-RA. Arch Pathol Lab Med. 2014. PMID: 25357115 Review.
Cited by
-
Deep learning-based histopathological assessment of tubulo-interstitial injury in chronic kidney diseases.Commun Med (Lond). 2025 Jan 5;5(1):3. doi: 10.1038/s43856-024-00708-3. Commun Med (Lond). 2025. PMID: 39757253 Free PMC article.
-
Metformin Alleviates Doxorubicin-Induced Cardiotoxicity via Preserving Mitochondrial Dynamics Balance and Calcium Homeostasis.Appl Biochem Biotechnol. 2025 Apr;197(4):2713-2733. doi: 10.1007/s12010-024-05141-9. Epub 2025 Jan 10. Appl Biochem Biotechnol. 2025. PMID: 39792339 Free PMC article.
-
The impact of consuming different types of high-caloric fat diet on the metabolic status, liver, and aortic integrity in rats.Sci Rep. 2024 Aug 10;14(1):18602. doi: 10.1038/s41598-024-68299-6. Sci Rep. 2024. PMID: 39127712 Free PMC article.
-
Refractive Index Changes of Cells and Cellular Compartments Upon Paraformaldehyde Fixation Acquired by Tomographic Phase Microscopy.Cytometry A. 2021 Apr;99(4):388-398. doi: 10.1002/cyto.a.24229. Epub 2020 Oct 19. Cytometry A. 2021. PMID: 32959478 Free PMC article.
-
The Application of Guanidinium to Improve Biomolecule Quality in Fixed, Paraffin-embedded Tissue.J Histochem Cytochem. 2023 Feb;71(2):87-101. doi: 10.1369/00221554231159451. Epub 2023 Mar 4. J Histochem Cytochem. 2023. PMID: 36869703 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

