Whole genome sequencing for deciphering the resistome of Chryseobacterium indologenes, an emerging multidrug-resistant bacterium isolated from a cystic fibrosis patient in Marseille, France
- PMID: 27222716
- PMCID: PMC4873609
- DOI: 10.1016/j.nmni.2016.03.006
Whole genome sequencing for deciphering the resistome of Chryseobacterium indologenes, an emerging multidrug-resistant bacterium isolated from a cystic fibrosis patient in Marseille, France
Abstract
We decipher the resistome of Chryseobacterium indologenes MARS15, an emerging multidrug-resistant clinical strain, using the whole genome sequencing strategy. The bacterium was isolated from the sputum of a hospitalized patient with cystic fibrosis in the Timone Hospital in Marseille, France. Genome sequencing was done with Illumina MiSeq using a paired-end strategy. The in silico analysis was done by RAST, the resistome by the ARG-ANNOT database and detection of polyketide synthase (PKS) by ANTISMAH. The genome size of C. indologenes MARS15 is 4 972 580 bp with 36.4% GC content. This multidrug-resistant bacterium was resistant to all β-lactams, including imipenem, and also to colistin. The resistome of C. indologenes MARS15 includes Ambler class A and B β-lactams encoding bla CIA and bla IND-2 genes and MBL (metallo-β-lactamase) genes, the CAT (chloramphenicol acetyltransferase) gene and the multidrug efflux pump AcrB. Specific features include the presence of an urease operon, an intact prophage and a carotenoid biosynthesis pathway. Interestingly, we report for the first time in C. indologenes a PKS cluster that might be responsible for secondary metabolite biosynthesis, similar to erythromycin. The whole genome sequence analysis provides insight into the resistome and the discovery of new details, such as the PKS cluster.
Keywords: Carotenoid; Chryseobacterium indologenes; microbial genomics; nonribosomal polyketide synthase; polyketide synthase; resistome; urease.
Figures


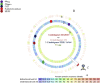
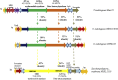
References
-
- Vandamme P., Bernardet J.F., Segers P., Kersters K., Holmes B. New perspectives in the classification of the flavobacteria: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int J Syst Bacteriol. 1994;44:827–831.
-
- Deng L., Li M.F., Li Y.H., Yang J.L., Zhou X. Chryseobacterium indologenes in four patients with leukemia. Transpl Infect Dis. 2015;17:583–587. - PubMed
-
- Chen X.Y., Zhao R., Chen Z.L., Liu L., Li X.D., Li Y.H. Chryseobacterium polytrichastri sp. nov., isolated from a moss (Polytrichastrum formosum), and emended description of the genus Chryseobacterium. Antonie Van Leeuwenhoek. 2015;107:403–410. - PubMed
-
- Chen F.L., Wang G.C., Teng S.O., Ou T.Y., Yu F.L., Lee W.S. Clinical and epidemiological features of Chryseobacterium indologenes infections: analysis of 215 cases. J Microbiol Immunol Infect. 2013;46:425–432. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
