The Type VI Secretion System in Escherichia coli and Related Species
- PMID: 27223818
- PMCID: PMC11575709
- DOI: 10.1128/ecosalplus.ESP-0009-2015
The Type VI Secretion System in Escherichia coli and Related Species
Abstract
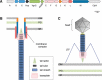
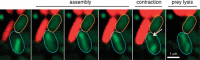
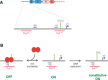
The type VI secretion system (T6SS) is a multiprotein complex widespread in Proteobacteria and dedicated to the delivery of toxins into both prokaryotic and eukaryotic cells. It thus participates in interbacterial competition as well as pathogenesis. The T6SS is a contractile weapon, related to the injection apparatus of contractile tailed bacteriophages. Basically, it assembles an inner tube wrapped by a sheath-like structure and anchored to the cell envelope via a membrane complex. The energy released by the contraction of the sheath propels the inner tube through the membrane channel and toward the target cell. Although the assembly and the mechanism of action are conserved across species, the repertoire of secreted toxins and the diversity of the regulatory mechanisms and of target cells make the T6SS a highly versatile secretion system. The T6SS is particularly represented in Escherichia coli pathotypes and Salmonella serotypes. In this review we summarize the current knowledge regarding the prevalence, the assembly, the regulation, and the roles of the T6SS in E. coli, Salmonella, and related species.
Figures







Similar articles
-
Fur-Dam Regulatory Interplay at an Internal Promoter of the Enteroaggregative Escherichia coli Type VI Secretion sci1 Gene Cluster.J Bacteriol. 2020 Apr 27;202(10):e00075-20. doi: 10.1128/JB.00075-20. Print 2020 Apr 27. J Bacteriol. 2020. PMID: 32152218 Free PMC article.
-
Role and Recruitment of the TagL Peptidoglycan-Binding Protein during Type VI Secretion System Biogenesis.J Bacteriol. 2019 May 22;201(12):e00173-19. doi: 10.1128/JB.00173-19. Print 2019 Jun 15. J Bacteriol. 2019. PMID: 30910811 Free PMC article.
-
The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization.PLoS Genet. 2015 Oct 13;11(10):e1005545. doi: 10.1371/journal.pgen.1005545. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26460929 Free PMC article.
-
TssA: The cap protein of the Type VI secretion system tail.Bioessays. 2017 Oct;39(10). doi: 10.1002/bies.201600262. Epub 2017 Aug 17. Bioessays. 2017. PMID: 28817192 Review.
-
Structure and Activity of the Type VI Secretion System.Microbiol Spectr. 2019 Jul;7(4):10.1128/microbiolspec.psib-0031-2019. doi: 10.1128/microbiolspec.PSIB-0031-2019. Microbiol Spectr. 2019. PMID: 31298206 Free PMC article. Review.
Cited by
-
In Silico Characterization of a Hypothetical Protein from Shigella dysenteriae ATCC 12039 Reveals a Pathogenesis-Related Protein of the Type-VI Secretion System.Bioinform Biol Insights. 2021 Apr 22;15:11779322211011140. doi: 10.1177/11779322211011140. eCollection 2021. Bioinform Biol Insights. 2021. PMID: 33994781 Free PMC article.
-
Microbial community composition interacts with local abiotic conditions to drive colonization resistance in human gut microbiome samples.Proc Biol Sci. 2021 Mar 31;288(1947):20203106. doi: 10.1098/rspb.2020.3106. Epub 2021 Mar 24. Proc Biol Sci. 2021. PMID: 33757361 Free PMC article.
-
Comparison of endogenous amino acid losses in broilers when offered nitrogen-free diets with differing ratios of dextrose to corn starch.Sci Rep. 2022 Apr 5;12(1):5689. doi: 10.1038/s41598-022-09746-0. Sci Rep. 2022. PMID: 35383258 Free PMC article.
-
Identification and Characterization of EvpQ, a Novel T6SS Effector Encoded on a Mobile Genetic Element in Edwardsiella piscicida.Front Microbiol. 2021 Mar 11;12:643498. doi: 10.3389/fmicb.2021.643498. eCollection 2021. Front Microbiol. 2021. PMID: 33776977 Free PMC article.
-
Comparative genome analyses of five Vibrio penaeicida strains provide insights into their virulence-related factors.Microb Genom. 2022 Feb;8(2):000766. doi: 10.1099/mgen.0.000766. Microb Genom. 2022. PMID: 35171089 Free PMC article.
References
-
- Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. 10.1186/1471-2164-10-104. [PubMed]10.1186/1471-2164-10-104 - DOI - DOI - PMC - PubMed
-
- Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. 2014. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16:227–236. [PubMed]10.1016/j.chom.2014.07.007 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

