Systems Metabolic Engineering of Escherichia coli
- PMID: 27223822
- PMCID: PMC11575710
- DOI: 10.1128/ecosalplus.ESP-0010-2015
Systems Metabolic Engineering of Escherichia coli
Abstract
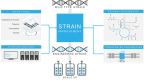
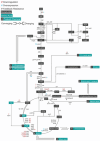
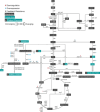
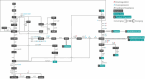
Systems metabolic engineering, which recently emerged as metabolic engineering integrated with systems biology, synthetic biology, and evolutionary engineering, allows engineering of microorganisms on a systemic level for the production of valuable chemicals far beyond its native capabilities. Here, we review the strategies for systems metabolic engineering and particularly its applications in Escherichia coli. First, we cover the various tools developed for genetic manipulation in E. coli to increase the production titers of desired chemicals. Next, we detail the strategies for systems metabolic engineering in E. coli, covering the engineering of the native metabolism, the expansion of metabolism with synthetic pathways, and the process engineering aspects undertaken to achieve higher production titers of desired chemicals. Finally, we examine a couple of notable products as case studies produced in E. coli strains developed by systems metabolic engineering. The large portfolio of chemical products successfully produced by engineered E. coli listed here demonstrates the sheer capacity of what can be envisioned and achieved with respect to microbial production of chemicals. Systems metabolic engineering is no longer in its infancy; it is now widely employed and is also positioned to further embrace next-generation interdisciplinary principles and innovation for its upgrade. Systems metabolic engineering will play increasingly important roles in developing industrial strains including E. coli that are capable of efficiently producing natural and nonnatural chemicals and materials from renewable nonfood biomass.
Figures



Similar articles
-
Systems Metabolic Engineering of Escherichia coli.EcoSal Plus. 2017 Mar;7(2). doi: 10.1128/ecosalplus.ESP-0088-2015. EcoSal Plus. 2017. PMID: 28281437 Review.
-
Construction and optimization of synthetic pathways in metabolic engineering.Curr Opin Microbiol. 2010 Jun;13(3):363-70. doi: 10.1016/j.mib.2010.02.004. Epub 2010 Mar 10. Curr Opin Microbiol. 2010. PMID: 20219419 Review.
-
Escherichia coli as a platform microbial host for systems metabolic engineering.Essays Biochem. 2021 Jul 26;65(2):225-246. doi: 10.1042/EBC20200172. Essays Biochem. 2021. PMID: 33956149 Review.
-
Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology.J Biomed Biotechnol. 2010;2010:761042. doi: 10.1155/2010/761042. Epub 2010 Apr 6. J Biomed Biotechnol. 2010. PMID: 20414363 Free PMC article. Review.
-
Systems metabolic engineering design: fatty acid production as an emerging case study.Biotechnol Bioeng. 2014 May;111(5):849-57. doi: 10.1002/bit.25205. Epub 2014 Feb 24. Biotechnol Bioeng. 2014. PMID: 24481660 Free PMC article. Review.
Cited by
-
Engineering the xylose-catabolizing Dahms pathway for production of poly(d-lactate-co-glycolate) and poly(d-lactate-co-glycolate-co-d-2-hydroxybutyrate) in Escherichia coli.Microb Biotechnol. 2017 Nov;10(6):1353-1364. doi: 10.1111/1751-7915.12721. Epub 2017 Apr 19. Microb Biotechnol. 2017. PMID: 28425205 Free PMC article.
-
Optimizing Escherichia coli strains and fermentation processes for enhanced L-lysine production: a review.Front Microbiol. 2024 Oct 4;15:1485624. doi: 10.3389/fmicb.2024.1485624. eCollection 2024. Front Microbiol. 2024. PMID: 39430105 Free PMC article. Review.
-
Engineering Clostridial Aldehyde/Alcohol Dehydrogenase for Selective Butanol Production.mBio. 2019 Jan 22;10(1):e02683-18. doi: 10.1128/mBio.02683-18. mBio. 2019. PMID: 30670620 Free PMC article.
-
Application of different types of CRISPR/Cas-based systems in bacteria.Microb Cell Fact. 2020 Sep 3;19(1):172. doi: 10.1186/s12934-020-01431-z. Microb Cell Fact. 2020. PMID: 32883277 Free PMC article. Review.
-
Microbial production of vitamin B12: a review and future perspectives.Microb Cell Fact. 2017 Jan 30;16(1):15. doi: 10.1186/s12934-017-0631-y. Microb Cell Fact. 2017. PMID: 28137297 Free PMC article. Review.
References
-
- Ahmed SA, Awosika J, Baldwin C, Bishop-Lilly KA, Biswas B, Broomall S, Chain PS, Chertkov O, Chokoshvili O, Coyne S, Davenport K, Detter JC, Dorman W, Erkkila TH, Folster JP, Frey KG, George M, Gleasner C, Henry M, Hill KK, Hubbard K, Insalaco J, Johnson S, Kitzmiller A, Krepps M, Lo CC, Luu T, McNew LA, Minogue T, Munk CA, Osborne B, Patel M, Reitenga KG, Rosenzweig CN, Shea A, Shen X, Strockbine N, Tarr C, Teshima H, van Gieson E, Verratti K, Wolcott M, Xie G, Sozhamannan S, Gibbons HS, Threat Characterization Consortium. 2012. Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including Shiga toxin encoding phage Stx2. PLoS One 7:e48228. 10.1371/journal.pone.0048228. 10.1371/journal.pone.0048228 - DOI - DOI - PMC - PubMed
-
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. [PubMed]10.1126/science.277.5331.1453 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

