SDCCAG8 Interacts with RAB Effector Proteins RABEP2 and ERC1 and Is Required for Hedgehog Signaling
- PMID: 27224062
- PMCID: PMC4880186
- DOI: 10.1371/journal.pone.0156081
SDCCAG8 Interacts with RAB Effector Proteins RABEP2 and ERC1 and Is Required for Hedgehog Signaling
Abstract
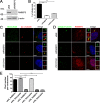
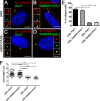
Recessive mutations in the SDCCAG8 gene cause a nephronophthisis-related ciliopathy with Bardet-Biedl syndrome-like features in humans. Our previous characterization of the orthologous Sdccag8gt/gt mouse model recapitulated the retinal-renal disease phenotypes and identified impaired DNA damage response signaling as an underlying disease mechanism in the kidney. However, several other phenotypic and mechanistic features of Sdccag8gt/gt mice remained unexplored. Here we show that Sdccag8gt/gt mice exhibit developmental and structural abnormalities of the skeleton and limbs, suggesting impaired Hedgehog (Hh) signaling. Indeed, cell culture studies demonstrate the requirement of SDCCAG8 for ciliogenesis and Hh signaling. Using an affinity proteomics approach, we demonstrate that SDCCAG8 interacts with proteins of the centriolar satellites (OFD1, AZI1), of the endosomal sorting complex (RABEP2, ERC1), and with non-muscle myosin motor proteins (MYH9, MYH10, MYH14) at the centrosome. Furthermore, we show that RABEP2 localization at the centrosome is regulated by SDCCAG8. siRNA mediated RABEP2 knockdown in hTERT-RPE1 cells leads to defective ciliogenesis, indicating a critical role for RABEP2 in this process. Together, this study identifies several centrosome-associated proteins as novel SDCCAG8 interaction partners, and provides new insights into the function of SDCCAG8 at this structure.
Conflict of interest statement
Figures




References
-
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426(6962):83–7. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

