Screen-identified selective inhibitor of lysine demethylase 5A blocks cancer cell growth and drug resistance
- PMID: 27224921
- PMCID: PMC5129982
- DOI: 10.18632/oncotarget.9539
Screen-identified selective inhibitor of lysine demethylase 5A blocks cancer cell growth and drug resistance
Abstract
Lysine demethylase 5A (KDM5A/RBP2/JARID1A) is a histone lysine demethylase that is overexpressed in several human cancers including lung, gastric, breast and liver cancers. It plays key roles in important cancer processes including tumorigenesis, metastasis, and drug tolerance, making it a potential cancer therapeutic target. Chemical tools to analyze KDM5A demethylase activity are extremely limited as available inhibitors are not specific for KDM5A. Here, we characterized KDM5A using a homogeneous luminescence-based assay and conducted a screen of about 9,000 small molecules for inhibitors. From this screen, we identified several 3-thio-1,2,4-triazole compounds that inhibited KDM5A with low μM in vitro IC50 values. Importantly, these compounds showed great specificity and did not inhibit its close homologue KDM5B (PLU1/JARID1B) or the related H3K27 demethylases KDM6A (UTX) and KDM6B (JMJD3). One compound, named YUKA1, was able to increase H3K4me3 levels in human cells and selectively inhibit the proliferation of cancer cells whose growth depends on KDM5A. As KDM5A was shown to mediate drug tolerance, we investigated the ability of YUKA1 to prevent drug tolerance in EGFR-mutant lung cancer cells treated with gefitinib and HER2+ breast cancer cells treated with trastuzumab. Remarkably, this compound hindered the emergence of drug-tolerant cells, highlighting the critical role of KDM5A demethylase activity in drug resistance. The small molecules presented here are excellent tool compounds for further study of KDM5A's demethylase activity and its contributions to cancer.
Keywords: KDM5A; anti-cancer drug; drug resistance; epigenetics; histone demethylase inhibitor.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the content of this article.
Figures

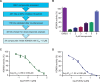
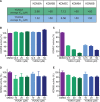

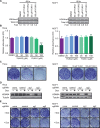
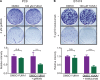
References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. - PubMed
-
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

