Genome-wide association study identifies 74 loci associated with educational attainment
- PMID: 27225129
- PMCID: PMC4883595
- DOI: 10.1038/nature17671
Genome-wide association study identifies 74 loci associated with educational attainment
Abstract
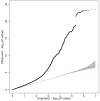
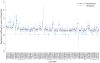
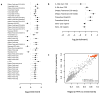
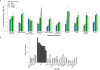
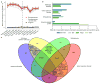
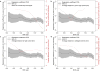
Educational attainment is strongly influenced by social and other environmental factors, but genetic factors are estimated to account for at least 20% of the variation across individuals. Here we report the results of a genome-wide association study (GWAS) for educational attainment that extends our earlier discovery sample of 101,069 individuals to 293,723 individuals, and a replication study in an independent sample of 111,349 individuals from the UK Biobank. We identify 74 genome-wide significant loci associated with the number of years of schooling completed. Single-nucleotide polymorphisms associated with educational attainment are disproportionately found in genomic regions regulating gene expression in the fetal brain. Candidate genes are preferentially expressed in neural tissue, especially during the prenatal period, and enriched for biological pathways involved in neural development. Our findings demonstrate that, even for a behavioural phenotype that is mostly environmentally determined, a well-powered GWAS identifies replicable associated genetic variants that suggest biologically relevant pathways. Because educational attainment is measured in large numbers of individuals, it will continue to be useful as a proxy phenotype in efforts to characterize the genetic influences of related phenotypes, including cognition and neuropsychiatric diseases.
Figures

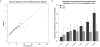





Comment in
-
Commentary: Genome-wide association study identifies 74 loci associated with educational attainment.Front Mol Neurosci. 2017 Jan 31;10:23. doi: 10.3389/fnmol.2017.00023. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28197077 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
- R01 DA036216/DA/NIDA NIH HHS/United States
- RF1 AG015819/AG/NIA NIH HHS/United States
- U01 AG009740/AG/NIA NIH HHS/United States
- MC_UU_12013/3/MRC_/Medical Research Council/United Kingdom
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- P2C HD047879/HD/NICHD NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- MC_UU_12013/1/MRC_/Medical Research Council/United Kingdom
- T32-AG000186-23/AG/NIA NIH HHS/United States
- P01 AG005842/AG/NIA NIH HHS/United States
- R37 DA005147/DA/NIDA NIH HHS/United States
- 647648/ERC_/European Research Council/International
- R01 AG017917/AG/NIA NIH HHS/United States
- MR/J012165/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12013/4/MRC_/Medical Research Council/United Kingdom
- CZD/16/6/4/CSO_/Chief Scientist Office/United Kingdom
- MC_PC_U127561128/MRC_/Medical Research Council/United Kingdom
- P01-AG005842/AG/NIA NIH HHS/United States
- R01-AG042568/AG/NIA NIH HHS/United States
- P30-AG012810/AG/NIA NIH HHS/United States
- P30 AG012810/AG/NIA NIH HHS/United States
- R01 AG042568/AG/NIA NIH HHS/United States
- T32 AG000186/AG/NIA NIH HHS/United States
- BB/F022441/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P01-AG005842-20S2/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

