Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development
- PMID: 27226300
- PMCID: PMC4988616
- DOI: 10.1073/pnas.1522811113
Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development
Abstract
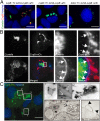
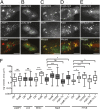
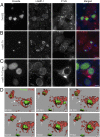
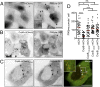
The Q fever bacterium Coxiella burnetii replicates inside host cells within a large Coxiella-containing vacuole (CCV) whose biogenesis relies on the Dot/Icm-dependent secretion of bacterial effectors. Several membrane trafficking pathways contribute membranes, proteins, and lipids for CCV biogenesis. These include the endocytic and autophagy pathways, which are characterized by phosphatidylinositol 3-phosphate [PI(3)P]-positive membranes. Here we show that the C. burnetii secreted effector Coxiella vacuolar protein B (CvpB) binds PI(3)P and phosphatidylserine (PS) on CCVs and early endosomal compartments and perturbs the activity of the phosphatidylinositol 5-kinase PIKfyve to manipulate PI(3)P metabolism. CvpB association to early endosome triggers vacuolation and clustering, leading to the channeling of large PI(3)P-positive membranes to CCVs for vacuole expansion. At CCVs, CvpB binding to early endosome- and autophagy-derived PI(3)P and the concomitant inhibition of PIKfyve favor the association of the autophagosomal machinery to CCVs for optimal homotypic fusion of the Coxiella-containing compartments. The importance of manipulating PI(3)P metabolism is highlighted by mutations in cvpB resulting in a multivacuolar phenotype, rescuable by gene complementation, indicative of a defect in CCV biogenesis. Using the insect model Galleria mellonella, we demonstrate the in vivo relevance of defective CCV biogenesis by highlighting an attenuated virulence phenotype associated with cvpB mutations.
Keywords: Coxiella burnetii; host–pathogen interactions; phosphoinositides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Madariaga MG, Rezai K, Trenholme GM, Weinstein RA. Q fever: A biological weapon in your backyard. Lancet Infect Dis. 2003;3(11):709–721. - PubMed
-
- Moffatt JH, Newton P, Newton HJ. Coxiella burnetii: Turning hostility into a home. Cell Microbiol. 2015;17(5):621–631. - PubMed
-
- Campoy EM, Mansilla ME, Colombo MI. Endocytic SNAREs are involved in optimal Coxiella burnetii vacuole development. Cell Microbiol. 2013;15(6):922–941. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

