Targeted Mutagenesis of Guinea Pig Cytomegalovirus Using CRISPR/Cas9-Mediated Gene Editing
- PMID: 27226370
- PMCID: PMC4944286
- DOI: 10.1128/JVI.00139-16
Targeted Mutagenesis of Guinea Pig Cytomegalovirus Using CRISPR/Cas9-Mediated Gene Editing
Abstract
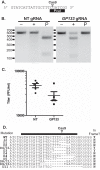
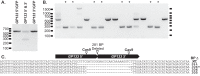
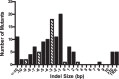
The cytomegaloviruses (CMVs) are among the most genetically complex mammalian viruses, with viral genomes that often exceed 230 kbp. Manipulation of cytomegalovirus genomes is largely performed using infectious bacterial artificial chromosomes (BACs), which necessitates the maintenance of the viral genome in Escherichia coli and successful reconstitution of virus from permissive cells after transfection of the BAC. Here we describe an alternative strategy for the mutagenesis of guinea pig cytomegalovirus that utilizes clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated genome editing to introduce targeted mutations to the viral genome. Transient transfection and drug selection were used to restrict lytic replication of guinea pig cytomegalovirus to cells that express Cas9 and virus-specific guide RNA. The result was highly efficient editing of the viral genome that introduced targeted insertion or deletion mutations to nonessential viral genes. Cotransfection of multiple virus-specific guide RNAs or a homology repair template was used for targeted, markerless deletions of viral sequence or to introduce exogenous sequence by homology-driven repair. As CRISPR/Cas9 mutagenesis occurs directly in infected cells, this methodology avoids selective pressures that may occur during propagation of the viral genome in bacteria and may facilitate genetic manipulation of low-passage or clinical CMV isolates.
Importance: The cytomegalovirus genome is complex, and viral adaptations to cell culture have complicated the study of infection in vivo Recombineering of viral bacterial artificial chromosomes enabled the study of recombinant cytomegaloviruses. Here we report the development of an alternative approach using CRISPR/Cas9-based mutagenesis in guinea pig cytomegalovirus, a small-animal model of congenital cytomegalovirus disease. CRISPR/Cas9 mutagenesis can introduce the same types of mutations to the viral genome as bacterial artificial chromosome recombineering but does so directly in virus-infected cells. CRISPR/Cas9 mutagenesis is not dependent on a bacterial intermediate, and defined viral mutants can be recovered after a limited number of viral genome replications, minimizing the risk of spontaneous mutation.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Repair of a Mutation Disrupting the Guinea Pig Cytomegalovirus Pentameric Complex Acquired during Fibroblast Passage Restores Pathogenesis in Immune-Suppressed Guinea Pigs and in the Context of Congenital Infection.J Virol. 2016 Aug 12;90(17):7715-27. doi: 10.1128/JVI.00320-16. Print 2016 Sep 1. J Virol. 2016. PMID: 27307567 Free PMC article.
-
Mutagenesis and Genome Engineering of Epstein-Barr Virus in Cultured Human Cells by CRISPR/Cas9.Methods Mol Biol. 2017;1498:23-31. doi: 10.1007/978-1-4939-6472-7_2. Methods Mol Biol. 2017. PMID: 27709566
-
Efficient Mutagenesis of Marek's Disease Virus-Encoded microRNAs Using a CRISPR/Cas9-Based Gene Editing System.Viruses. 2020 Apr 20;12(4):466. doi: 10.3390/v12040466. Viruses. 2020. PMID: 32325942 Free PMC article.
-
[The application of CRISPR-Cas9 gene editing technology in viral infection diseases].Yi Chuan. 2015 May;37(5):412-8. doi: 10.16288/j.yczz.14-460. Yi Chuan. 2015. PMID: 25998428 Review. Chinese.
-
Potential Application of the CRISPR/Cas9 System against Herpesvirus Infections.Viruses. 2018 May 29;10(6):291. doi: 10.3390/v10060291. Viruses. 2018. PMID: 29844277 Free PMC article. Review.
Cited by
-
Concurrent Gene Insertion, Deletion, and Inversion during the Construction of a Novel Attenuated BoHV-1 Using CRISPR/Cas9 Genome Editing.Vet Sci. 2022 Mar 30;9(4):166. doi: 10.3390/vetsci9040166. Vet Sci. 2022. PMID: 35448664 Free PMC article.
-
Simultaneous Deletion of Virulence Factors and Insertion of Antigens into the Infectious Laryngotracheitis Virus Using NHEJ-CRISPR/Cas9 and Cre-Lox System for Construction of a Stable Vaccine Vector.Vaccines (Basel). 2019 Dec 5;7(4):207. doi: 10.3390/vaccines7040207. Vaccines (Basel). 2019. PMID: 31817447 Free PMC article.
-
BIRC5 Gene Disruption via CRISPR/Cas9n Platform Suppress Acute Myelocytic Leukemia Progression.Iran Biomed J. 2019 Nov;23(6):369-78. doi: 10.29252/ibj.23.6.369. Epub 2019 May 20. Iran Biomed J. 2019. PMID: 31104397 Free PMC article.
-
V5 and GFP Tagging of Viral Gene pp38 of Marek's Disease Vaccine Strain CVI988 Using CRISPR/Cas9 Editing.Viruses. 2022 Feb 21;14(2):436. doi: 10.3390/v14020436. Viruses. 2022. PMID: 35216029 Free PMC article.
-
Construction of a highly efficient CRISPR/Cas9-mediated duck enteritis virus-based vaccine against H5N1 avian influenza virus and duck Tembusu virus infection.Sci Rep. 2017 May 3;7(1):1478. doi: 10.1038/s41598-017-01554-1. Sci Rep. 2017. PMID: 28469192 Free PMC article.
References
-
- Mocarski ES Jr, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA III, Kouzarides T, Martignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston KM, Barrell BG. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol 154:125–169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

