Biochemical Characterization of the Human Mitochondrial Replicative Twinkle Helicase: SUBSTRATE SPECIFICITY, DNA BRANCH MIGRATION, AND ABILITY TO OVERCOME BLOCKADES TO DNA UNWINDING
- PMID: 27226550
- PMCID: PMC4933186
- DOI: 10.1074/jbc.M115.712026
Biochemical Characterization of the Human Mitochondrial Replicative Twinkle Helicase: SUBSTRATE SPECIFICITY, DNA BRANCH MIGRATION, AND ABILITY TO OVERCOME BLOCKADES TO DNA UNWINDING
Abstract
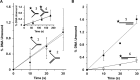
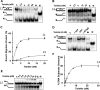
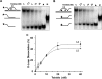
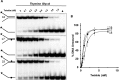
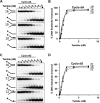
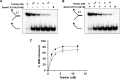
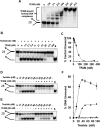
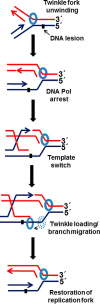
Mutations in the c10orf2 gene encoding the human mitochondrial DNA replicative helicase Twinkle are linked to several rare genetic diseases characterized by mitochondrial defects. In this study, we have examined the catalytic activity of Twinkle helicase on model replication fork and DNA repair structures. Although Twinkle behaves as a traditional 5' to 3' helicase on conventional forked duplex substrates, the enzyme efficiently dissociates D-loop DNA substrates irrespective of whether it possesses a 5' or 3' single-stranded tailed invading strand. In contrast, we report for the first time that Twinkle branch-migrates an open-ended mobile three-stranded DNA structure with a strong 5' to 3' directionality preference. To determine how well Twinkle handles potential roadblocks to mtDNA replication, we tested the ability of the helicase to unwind substrates with site-specific oxidative DNA lesions or bound by the mitochondrial transcription factor A. Twinkle helicase is inhibited by DNA damage in a unique manner that is dependent on the type of oxidative lesion and the strand in which it resides. Novel single molecule FRET binding and unwinding assays show an interaction of the excluded strand with Twinkle as well as events corresponding to stepwise unwinding and annealing. TFAM inhibits Twinkle unwinding, suggesting other replisome proteins may be required for efficient removal. These studies shed new insight on the catalytic functions of Twinkle on the key DNA structures it would encounter during replication or possibly repair of the mitochondrial genome and how well it tolerates potential roadblocks to DNA unwinding.
Keywords: DNA helicase; DNA repair; DNA replication; Twinkle; branch migration; genetic disease; helicase; mitochondria.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Shutt T. E., and Gray M. W. (2006) Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 62, 588–599 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

