Krüppel-like Factor 3 (KLF3/BKLF) Is Required for Widespread Repression of the Inflammatory Modulator Galectin-3 (Lgals3)
- PMID: 27226561
- PMCID: PMC4965555
- DOI: 10.1074/jbc.M116.715748
Krüppel-like Factor 3 (KLF3/BKLF) Is Required for Widespread Repression of the Inflammatory Modulator Galectin-3 (Lgals3)
Abstract
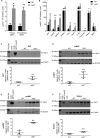
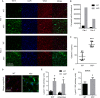
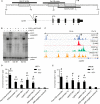
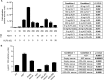
The Lgals3 gene encodes a multifunctional β-galactoside-binding protein, galectin-3. Galectin-3 has been implicated in a broad range of biological processes from chemotaxis and inflammation to fibrosis and apoptosis. The role of galectin-3 as a modulator of inflammation has been studied intensively, and recent evidence suggests that it may serve as a protective factor in obesity and other metabolic disorders. Despite considerable interest in galectin-3, little is known about its physiological regulation at the transcriptional level. Here, using knockout mice, chromatin immunoprecipitations, and cellular and molecular analyses, we show that the zinc finger transcription factor Krüppel-like factor 3 (KLF3) directly represses galectin-3 transcription. We find that galectin-3 is broadly up-regulated in KLF3-deficient mouse tissues, that KLF3 occupies regulatory regions of the Lgals3 gene, and that KLF3 directly binds its cognate elements (CACCC boxes) in the galectin-3 promoter and represses its activation in cellular assays. We also provide mechanistic insights into the regulation of Lgals3, demonstrating that C-terminal binding protein (CtBP) is required to drive optimal KLF3-mediated silencing. These findings help to enhance our understanding of how expression of the inflammatory modulator galectin-3 is controlled, opening up avenues for potential therapeutic interventions in the future.
Keywords: Krüppel-like factor (KLF); Krüppel-like factor 3 (KLF3); adipose tissue; galectin; galectin-3 (Lgals3); gene expression; gene regulation; inflammation; metabolism; transcription regulation.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- MacKinnon A. C., Farnworth S. L., Hodkinson P. S., Henderson N. C., Atkinson K. M., Leffler H., Nilsson U. J., Haslett C., Forbes S. J., and Sethi T. (2008) Regulation of alternative macrophage activation by galectin-3. J. Immunol. 180, 2650–2658 - PubMed
-
- Elad-Sfadia G., Haklai R., Balan E., and Kloog Y. (2004) Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J. Biol. Chem. 279, 34922–34930 - PubMed
-
- de Boer R. A., Voors A. A., Muntendam P., van Gilst W. H., and van Veldhuisen D. J. (2009) Galectin-3: a novel mediator of heart failure development and progression. Eur. J. Heart Fail. 11, 811–817 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

