Macrophage Migration Inhibitory Factor-CXCR4 Receptor Interactions: EVIDENCE FOR PARTIAL ALLOSTERIC AGONISM IN COMPARISON WITH CXCL12 CHEMOKINE
- PMID: 27226569
- PMCID: PMC4957068
- DOI: 10.1074/jbc.M116.717751
Macrophage Migration Inhibitory Factor-CXCR4 Receptor Interactions: EVIDENCE FOR PARTIAL ALLOSTERIC AGONISM IN COMPARISON WITH CXCL12 CHEMOKINE
Abstract
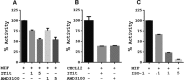
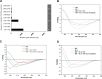
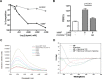
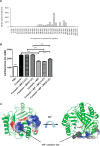
An emerging number of non-chemokine mediators are found to bind to classical chemokine receptors and to elicit critical biological responses. Macrophage migration inhibitory factor (MIF) is an inflammatory cytokine that exhibits chemokine-like activities through non-cognate interactions with the chemokine receptors CXCR2 and CXCR4, in addition to activating the type II receptor CD74. Activation of the MIF-CXCR2 and -CXCR4 axes promotes leukocyte recruitment, mediating the exacerbating role of MIF in atherosclerosis and contributing to the wealth of other MIF biological activities. Although the structural basis of the MIF-CXCR2 interaction has been well studied and was found to engage a pseudo-ELR and an N-like loop motif, nothing is known about the regions of CXCR4 and MIF that are involved in binding to each other. Using a genetic strain of Saccharomyces cerevisiae that expresses a functional CXCR4 receptor, site-specific mutagenesis, hybrid CXCR3/CXCR4 receptors, pharmacological reagents, peptide array analysis, chemotaxis, fluorescence spectroscopy, and circular dichroism, we provide novel molecular information about the structural elements that govern the interaction between MIF and CXCR4. The data identify similarities with classical chemokine-receptor interactions but also provide evidence for a partial allosteric agonist compared with CXCL12 that is possible due to the two binding sites of CXCR4.
Keywords: G protein-coupled receptor (GPCR); chemokine; cytokine; protein mapping; protein-protein interaction; structure-function.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Thelen M. (2001) Dancing to the tune of chemokines. Nat. Immunol. 2, 129–134 - PubMed
-
- Charo I. F., and Ransohoff R. M. (2006) The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 354, 610–621 - PubMed
-
- Moser B., and Loetscher P. (2001) Lymphocyte traffic control by chemokines. Nat. Immunol. 2, 123–128 - PubMed
-
- Bleul C. C., Farzan M., Choe H., Parolin C., Clark-Lewis I., Sodroski J., and Springer T. A. (1996) The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382, 829–833 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

