A Camelid-derived Antibody Fragment Targeting the Active Site of a Serine Protease Balances between Inhibitor and Substrate Behavior
- PMID: 27226628
- PMCID: PMC4946931
- DOI: 10.1074/jbc.M116.732503
A Camelid-derived Antibody Fragment Targeting the Active Site of a Serine Protease Balances between Inhibitor and Substrate Behavior
Abstract
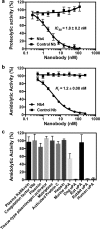
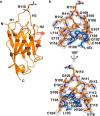
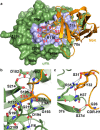
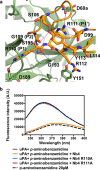
A peptide segment that binds the active site of a serine protease in a substrate-like manner may behave like an inhibitor or a substrate. However, there is sparse information on which factors determine the behavior a particular peptide segment will exhibit. Here, we describe the first x-ray crystal structure of a nanobody in complex with a serine protease. The nanobody displays a new type of interaction between an antibody and a serine protease as it inserts its complementary determining region-H3 loop into the active site of the protease in a substrate-like manner. The unique binding mechanism causes the nanobody to behave as a strong inhibitor as well as a poor substrate. Intriguingly, its substrate behavior is incomplete, as 30-40% of the nanobody remained intact and inhibitory after prolonged incubation with the protease. Biochemical analysis reveals that an intra-loop interaction network within the complementary determining region-H3 of the nanobody balances its inhibitor versus substrate behavior. Collectively, our results unveil molecular factors, which may be a general mechanism to determine the substrate versus inhibitor behavior of other protease inhibitors.
Keywords: biophysics; biotechnology; serine protease; single-domain antibody (sdAb, nanobody); x-ray crystallography.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

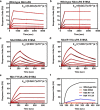
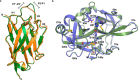


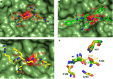
References
-
- Puente X, O., and López-Otín C. (2008) in The Cancer Degradome (Edwards D., Høyer-Hansen G., Blasi F., and Sloane B. F., eds) pp. 3–15, Springer, New York
-
- Turk B. (2006) Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 5, 785–799 - PubMed
-
- Schechter I., and Berger A. (1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

