Heterogeneity of Human Neutrophil CD177 Expression Results from CD177P1 Pseudogene Conversion
- PMID: 27227454
- PMCID: PMC4882059
- DOI: 10.1371/journal.pgen.1006067
Heterogeneity of Human Neutrophil CD177 Expression Results from CD177P1 Pseudogene Conversion
Abstract
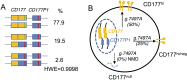
Most humans harbor both CD177neg and CD177pos neutrophils but 1-10% of people are CD177null, placing them at risk for formation of anti-neutrophil antibodies that can cause transfusion-related acute lung injury and neonatal alloimmune neutropenia. By deep sequencing the CD177 locus, we catalogued CD177 single nucleotide variants and identified a novel stop codon in CD177null individuals arising from a single base substitution in exon 7. This is not a mutation in CD177 itself, rather the CD177null phenotype arises when exon 7 of CD177 is supplied entirely by the CD177 pseudogene (CD177P1), which appears to have resulted from allelic gene conversion. In CD177 expressing individuals the CD177 locus contains both CD177P1 and CD177 sequences. The proportion of CD177hi neutrophils in the blood is a heritable trait. Abundance of CD177hi neutrophils correlates with homozygosity for CD177 reference allele, while heterozygosity for ectopic CD177P1 gene conversion correlates with increased CD177neg neutrophils, in which both CD177P1 partially incorporated allele and paired intact CD177 allele are transcribed. Human neutrophil heterogeneity for CD177 expression arises by ectopic allelic conversion. Resolution of the genetic basis of CD177null phenotype identifies a method for screening for individuals at risk of CD177 isoimmunisation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Stroncek DF. Neutrophil-specific antigen HNA-2a, NB1 glycoprotein, and CD177. Curr. Opin. Hematol. 2007;14(6):688–693. - PubMed
-
- Caruccio L, Bettinotti M, Director-Myska AE, Arthur DC, Stroncek D. The gene overexpressed in polycythemia rubra vera, PRV-1, and the gene encoding a neutrophil alloantigen, NB1, are alleles of a single gene, CD177, in chromosome band 19q13.31. Transfusion. 2006;46(3):441–447. - PubMed
-
- Skubitz KM, Stroncek DF, Sun B. Neutrophil-specific antigen NB1 is anchored via a glycosyl-phosphatidylinositol linkage. Journal of Leukocyte Biology. 1991;49(2):163–171. - PubMed
-
- Goldschmeding R, van Dalen CM, Gaber N, Calafat J, Huizinga TW, van der Schoot CE, et al. Further characterization of the NB 1 antigen as a variably expressed 56–62 kD GPI-linked glycoprotein of plasma membranes and specific granules of neutrophils. Br. J. Haematol. 1992;81(3):336–345. - PubMed
-
- Stroncek DF, Plachta LB, Herr GP, Dalmasso AP. Analysis of the expression of neutrophil-specific antigen NB1: characterization of neutrophils that react with but are not agglutinated by anti-NB1. Transfusion. 1993;33(8):656–660. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

