Genetic Characterization and Classification of Human and Animal Sapoviruses
- PMID: 27228126
- PMCID: PMC4881899
- DOI: 10.1371/journal.pone.0156373
Genetic Characterization and Classification of Human and Animal Sapoviruses
Abstract
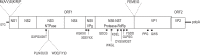
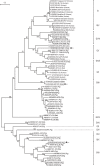
Sapoviruses (SaVs) are enteric caliciviruses that have been detected in multiple mammalian species, including humans, pigs, mink, dogs, sea lions, chimpanzees, and rats. They show a high level of diversity. A SaV genome commonly encodes seven nonstructural proteins (NSs), including the RNA polymerase protein NS7, and two structural proteins (VP1 and VP2). We classified human and animal SaVs into 15 genogroups (G) based on available VP1 sequences, including three newly characterized genomes from this study. We sequenced the full length genomes of one new genogroup V (GV), one GVII and one GVIII porcine SaV using long range RT-PCR including newly designed forward primers located in the conserved motifs of the putative NS3, and also 5' RACE methods. We also determined the 5'- and 3'-ends of sea lion GV SaV and canine GXIII SaV. Although the complete genomic sequences of GIX-GXII, and GXV SaVs are unavailable, common features of SaV genomes include: 1) "GTG" at the 5'-end of the genome, and a short (9~14 nt) 5'-untranslated region; and 2) the first five amino acids (M [A/V] S [K/R] P) of the putative NS1 and the five amino acids (FEMEG) surrounding the putative cleavage site between NS7 and VP1 were conserved among the chimpanzee, two of five genogroups of pig (GV and GVIII), sea lion, canine, and human SaVs. In contrast, these two amino acid motifs were clearly different in three genogroups of porcine (GIII, GVI and GVII), and bat SaVs. Our results suggest that several animal SaVs have genetic similarities to human SaVs. However, the ability of SaVs to be transmitted between humans and animals is uncertain.
Conflict of interest statement
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

