Improved Synthesis and In Vitro Evaluation of an Aptamer Ribosomal Toxin Conjugate
- PMID: 27228412
- PMCID: PMC4932783
- DOI: 10.1089/nat.2015.0599
Improved Synthesis and In Vitro Evaluation of an Aptamer Ribosomal Toxin Conjugate
Abstract
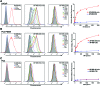
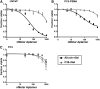
Delivery of toxins, such as the ricin A chain, Pseudomonas exotoxin, and gelonin, using antibodies has had some success in inducing specific toxicity in cancer treatments. However, these antibody-toxin conjugates, called immunotoxins, can be bulky, difficult to express, and may induce an immune response upon in vivo administration. We previously reported delivery of a recombinant variant of gelonin (rGel) by the full-length prostate-specific membrane antigen (PSMA) binding aptamer, A9, to potentially circumvent some of these problems. Here, we report a streamlined approach to generating aptamer-rGel conjugates utilizing a chemically synthesized minimized form of the A9 aptamer. Unlike the full-length A9 aptamer, this minimized variant can be chemically synthesized with a 5' terminal thiol. This facilitates the large scale synthesis and generation of aptamer toxin conjugates linked by a reducible disulfide linkage. Using this approach, we generated aptamer-toxin conjugates and evaluated their binding specificity and toxicity. On PSMA(+) LNCaP prostate cancer cells, the A9.min-rGel conjugate demonstrated an IC50 of ∼60 nM. Additionally, we performed a stability analysis of this conjugate in mouse serum where the conjugate displayed a t1/2 of ∼4 h, paving the way for future in vivo experiments.
Figures


References
-
- Pastan I, Hassan R, FitzGerald DJ. and Kreitman RJ. (2007). Immunotoxin treatment of cancer. Annu Rev Med 58:221–237 - PubMed
-
- Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA. and Langer R. (2004). Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res 64:7668–7672 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

