The essential genome of Streptococcus agalactiae
- PMID: 27229469
- PMCID: PMC4881062
- DOI: 10.1186/s12864-016-2741-z
The essential genome of Streptococcus agalactiae
Abstract
Background: Next-generation sequencing of transposon-genome junctions from a saturated bacterial mutant library (Tn-seq) is a powerful tool that permits genome-wide determination of the contribution of genes to fitness of the organism under a wide range of experimental conditions. We report development, testing, and results from a Tn-seq system for use in Streptococcus agalactiae (group B Streptococcus; GBS), an important cause of neonatal sepsis.
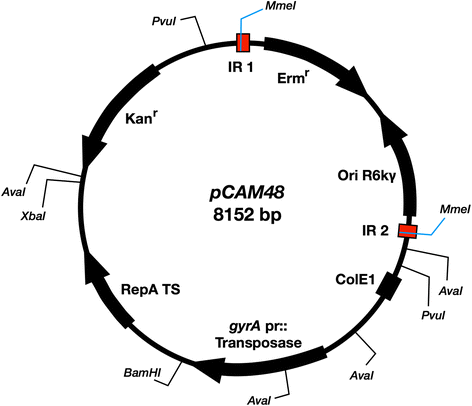
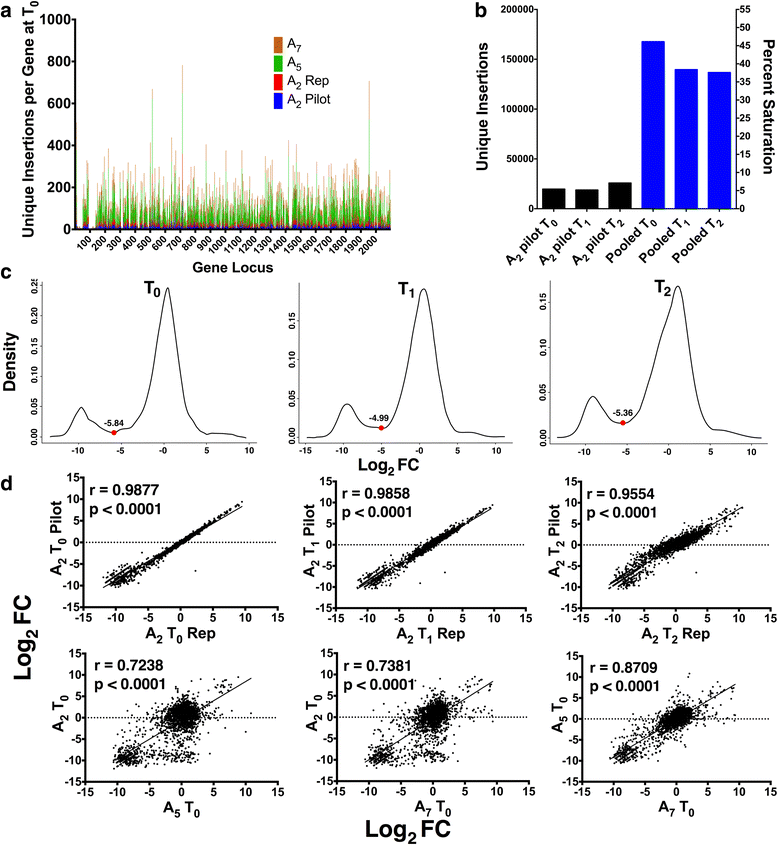
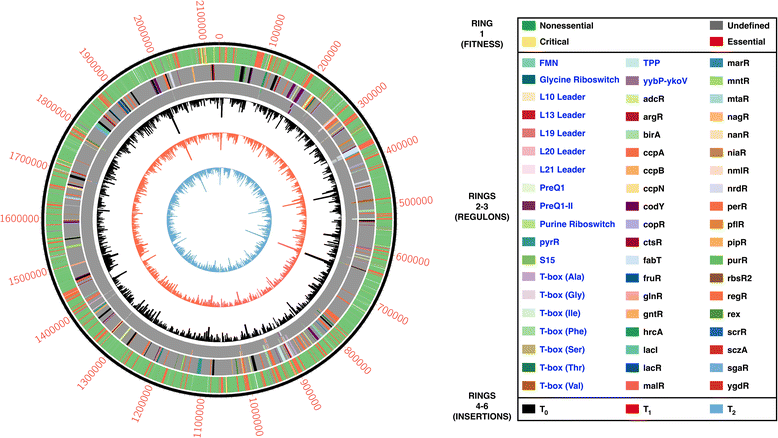
Methods: Our method uses a Himar1 mini-transposon that inserts at genomic TA dinucleotide sites, delivered to GBS on a temperature-sensitive plasmid that is subsequently cured from the bacterial population. In order to establish the GBS essential genome, we performed Tn-seq on DNA collected from three independent mutant libraries-with at least 135,000 mutants per library-at serial 24 h time points after outgrowth in rich media.
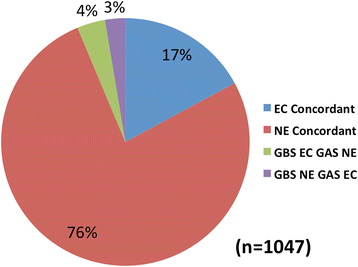
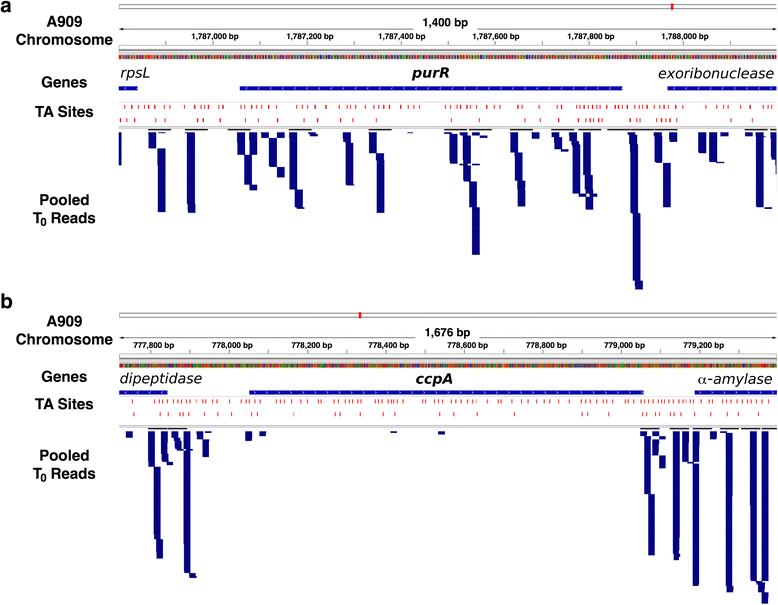
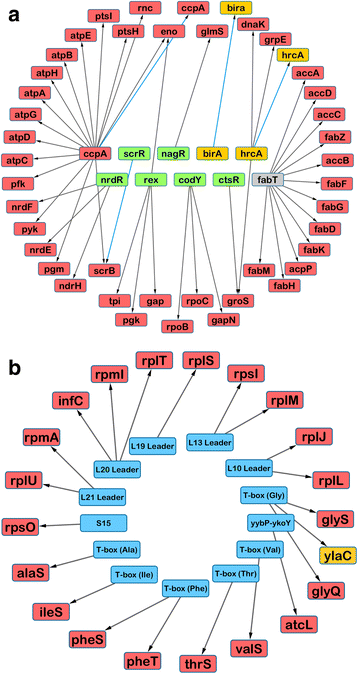
Results: After statistical analysis of transposon insertion density and distribution, we identified 13.5 % of genes as essential and 1.2 % as critical, with high levels of reproducibility. Essential and critical genes are enriched for fundamental cellular housekeeping functions, such as acyl-tRNA biosynthesis, nucleotide metabolism, and glycolysis. We further validated our system by comparing fitness assignments of homologous genes in GBS and a close bacterial relative, Streptococcus pyogenes, which demonstrated 93 % concordance. Finally, we used our fitness assignments to identify signal transduction pathway components predicted to be essential or critical in GBS.
Conclusions: We believe that our baseline fitness assignments will be a valuable tool for GBS researchers and that our system has the potential to reveal key pathogenesis gene networks and potential therapeutic/preventative targets.
Figures
References
-
- Campbell JR, Hillier SL, Krohn MA, Ferrieri P, Zaleznik DF, Baker CJ. Group B streptococcal colonization and serotype-specific immunity in pregnant women at delivery. Obstet Gynecol. 2000;96:498–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

