Innate immune responses in human hepatocyte-derived cell lines alter genotype 1 hepatitis E virus replication efficiencies
- PMID: 27230536
- PMCID: PMC4882509
- DOI: 10.1038/srep26827
Innate immune responses in human hepatocyte-derived cell lines alter genotype 1 hepatitis E virus replication efficiencies
Abstract
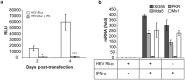
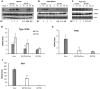
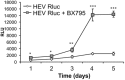
Hepatitis E virus (HEV) is a significant health problem in developing countries causing sporadic and epidemic forms of acute viral hepatitis. Hepatitis E is a self-limiting disease; however, chronic HEV infections are being reported in immunocompromised individuals. The disease severity is more during pregnancy with high mortality (20-25%), especially in third trimester. Early cellular responses after HEV infection are not completely understood. We analyzed innate immune responses associated with genotype-I HEV replication in human hepatoma cell lines (Huh7, Huh7.5 and HepG2/C3A) using HEV replicon system. These cells supported HEV replication with different efficiencies due to the cell type specific innate immune responses. HepG2/C3A cells were less supportive to HEV replication as compared to Huh7.5 and S10-3 cells. Reconstitution of the defective RIG-I and TLR3 signaling in Huh7.5 cells enabled them to induce higher level antiviral responses and restrict HEV replication, suggesting the involvement of both RIG-I and TLR3 in sensing HEV RNA and downstream activation of interferon regulatory factor 3 (IRF3) to generate antiviral responses. Inhibition of IRF3 mediated downstream responses in HepG2/C3A cells by pharmacological inhibitor BX795 significantly improved HEV replication efficiency implying the importance of this study in establishing a better cell culture system for future HEV studies.
Figures
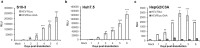

Similar articles
-
ISG15 Modulates Type I Interferon Signaling and the Antiviral Response during Hepatitis E Virus Replication.J Virol. 2017 Sep 12;91(19):e00621-17. doi: 10.1128/JVI.00621-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28724761 Free PMC article.
-
RIG-I is a key antiviral interferon-stimulated gene against hepatitis E virus regardless of interferon production.Hepatology. 2017 Jun;65(6):1823-1839. doi: 10.1002/hep.29105. Epub 2017 May 3. Hepatology. 2017. PMID: 28195391
-
Progesterone-Mediated Enhancement of Hepatitis E Virus Replication in Human Liver Cells.mBio. 2021 Jun 29;12(3):e0143421. doi: 10.1128/mBio.01434-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154410 Free PMC article.
-
Interplay between Hepatitis E Virus and Host Cell Pattern Recognition Receptors.Int J Mol Sci. 2021 Aug 26;22(17):9259. doi: 10.3390/ijms22179259. Int J Mol Sci. 2021. PMID: 34502167 Free PMC article. Review.
-
Stem Cell-Derived Culture Models of Hepatitis E Virus Infection.Cold Spring Harb Perspect Med. 2019 Mar 1;9(3):a031799. doi: 10.1101/cshperspect.a031799. Cold Spring Harb Perspect Med. 2019. PMID: 29686039 Free PMC article. Review.
Cited by
-
Molecular Biology and Infection of Hepatitis E Virus.Front Microbiol. 2016 Sep 7;7:1419. doi: 10.3389/fmicb.2016.01419. eCollection 2016. Front Microbiol. 2016. PMID: 27656178 Free PMC article. Review.
-
Multiple mechanisms drive phage infection efficiency in nearly identical hosts.ISME J. 2018 Jun;12(6):1605-1618. doi: 10.1038/s41396-018-0099-8. Epub 2018 Mar 22. ISME J. 2018. PMID: 29568113 Free PMC article.
-
Pan-Genotype Hepatitis E Virus Replication in Stem Cell-Derived Hepatocellular Systems.Gastroenterology. 2018 Feb;154(3):663-674.e7. doi: 10.1053/j.gastro.2017.10.041. Epub 2017 Dec 24. Gastroenterology. 2018. PMID: 29277559 Free PMC article.
-
Janus sword actions of chloroquine and hydroxychloroquine against COVID-19.Cell Signal. 2020 Sep;73:109706. doi: 10.1016/j.cellsig.2020.109706. Epub 2020 Jul 3. Cell Signal. 2020. PMID: 32629149 Free PMC article.
-
Hepatitis E Virus (HEV) egress: Role of BST2 (Tetherin) and interferon induced long non- coding RNA (lncRNA) BISPR.PLoS One. 2017 Nov 1;12(11):e0187334. doi: 10.1371/journal.pone.0187334. eCollection 2017. PLoS One. 2017. PMID: 29091957 Free PMC article.
References
-
- Holla R. P., Ahmad I., Ahmad Z. & Jameel S. Molecular virology of hepatitis E virus. Semin Liver Dis 33, 3–14 (2013). - PubMed
-
- Perttila J., Spuul P. & Ahola T. Early secretory pathway localization and lack of processing for hepatitis E virus replication protein pORF1. J Gen Virol 94, 807–816 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

