The Nim1 kinase Gin4 has distinct domains crucial for septin assembly, phospholipid binding and mitotic exit
- PMID: 27231094
- PMCID: PMC4958294
- DOI: 10.1242/jcs.183160
The Nim1 kinase Gin4 has distinct domains crucial for septin assembly, phospholipid binding and mitotic exit
Abstract
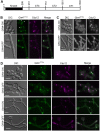
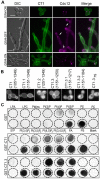
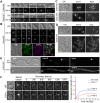
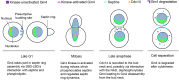
In fungi, the Nim1 protein kinases, such as Gin4, are important regulators of multiple cell cycle events, including the G2-M transition, septin assembly, polarized growth and cytokinesis. Compelling evidence has linked some key functions of Gin4 with the large C-terminal non-kinase region which, however, is poorly defined. By systematically dissecting and functionally characterizing the non-kinase region of Gin4 in the human fungal pathogen Candida albicans, we report the identification of three new domains with distinct functions: a lipid-binding domain (LBD), a septin-binding domain (SBD) and a nucleolus-associating domain (NAD). The LBD and SBD are indispensable for the function of Gin4, and they alone could sufficiently restore septin ring assembly in GIN4-null mutants. The NAD localizes to the periphery of the nucleolus and physically associates with Cdc14, the ultimate effector of the mitotic exit network. Gin4 mutants that lack the NAD are defective in spindle orientation and exit mitosis prematurely. Furthermore, we show that Gin4 is a substrate of Cdc14. These findings provide novel insights into the roles and mechanisms of Nim1 kinases in the regulation of some crucial cell cycle events.
Keywords: Candida albicans; Cdc14; Gin4; Nim1 kinases; Septin assembly.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

