Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
- PMID: 27231346
- PMCID: PMC4957043
- DOI: 10.1074/jbc.M115.711226
Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
Abstract
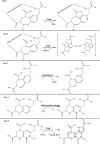
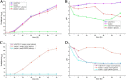
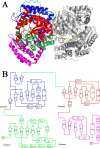
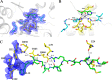
Pyrroloquinoline quinone (PQQ) has received considerable attention due to its numerous important physiological functions. PqqA is a precursor peptide of PQQ with two conserved residues: glutamate and tyrosine. After linkage of the Cγ of glutamate and Cϵ of tyrosine by PqqE, these two residues are hypothesized to be cleaved from PqqA by PqqF. The linked glutamate and tyrosine residues are then used to synthesize PQQ. Here, we demonstrated that the pqqF gene is essential for PQQ biosynthesis as deletion of it eliminated the inhibition of prodigiosin production by glucose. We further determined the crystal structure of PqqF, which has a closed clamshell-like shape. The PqqF consists of two halves composed of an N- and a C-terminal lobe. The PqqF-N and PqqF-C lobes form a chamber with the volume of the cavity of ∼9400 Å(3) The PqqF structure conforms to the general structure of inverzincins. Compared with the most thoroughly characterized inverzincin insulin-degrading enzyme, the size of PqqF chamber is markedly smaller, which may define the specificity for its substrate PqqA. Furthermore, the 14-amino acid-residue-long tag formed by the N-terminal tag from expression vector precisely protrudes into the counterpart active site; this N-terminal tag occupies the active site and stabilizes the closed, inactive conformation. His-48, His-52, Glu-129 and His-14 from the N-terminal tag coordinate with the zinc ion. Glu-51 acts as a base catalyst. The observed histidine residue-mediated inhibition may be applicable for the design of a peptide for the inhibition of M16 metalloproteases.
Keywords: crystal structure; enzyme inhibitor; metalloprotease; quinone; structure-function.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
The pyrroloquinoline quinone biosynthesis pathway revisited: a structural approach.BMC Biochem. 2008 Mar 27;9:8. doi: 10.1186/1471-2091-9-8. BMC Biochem. 2008. PMID: 18371220 Free PMC article.
-
Biogenesis of the peptide-derived redox cofactor pyrroloquinoline quinone.Curr Opin Chem Biol. 2020 Dec;59:93-103. doi: 10.1016/j.cbpa.2020.05.001. Epub 2020 Jul 27. Curr Opin Chem Biol. 2020. PMID: 32731194 Free PMC article. Review.
-
(1)H, (13)C, and (15)N resonance assignments and secondary structure information for Methylobacterium extorquens PqqD and the complex of PqqD with PqqA.Biomol NMR Assign. 2016 Oct;10(2):385-9. doi: 10.1007/s12104-016-9705-8. Epub 2016 Sep 16. Biomol NMR Assign. 2016. PMID: 27638737 Free PMC article.
-
A two-component protease in Methylorubrum extorquens with high activity toward the peptide precursor of the redox cofactor pyrroloquinoline quinone.J Biol Chem. 2019 Oct 11;294(41):15025-15036. doi: 10.1074/jbc.RA119.009684. Epub 2019 Aug 19. J Biol Chem. 2019. PMID: 31427437 Free PMC article.
-
Microbial synthesis of pyrroloquinoline quinone.World J Microbiol Biotechnol. 2023 Dec 7;40(1):31. doi: 10.1007/s11274-023-03833-8. World J Microbiol Biotechnol. 2023. PMID: 38057682 Review.
Cited by
-
Protease-associated import systems are widespread in Gram-negative bacteria.PLoS Genet. 2019 Oct 15;15(10):e1008435. doi: 10.1371/journal.pgen.1008435. eCollection 2019 Oct. PLoS Genet. 2019. PMID: 31613892 Free PMC article.
-
Occurrence, function, and biosynthesis of mycofactocin.Appl Microbiol Biotechnol. 2019 Apr;103(7):2903-2912. doi: 10.1007/s00253-019-09684-4. Epub 2019 Feb 18. Appl Microbiol Biotechnol. 2019. PMID: 30778644 Free PMC article. Review.
-
Characterization of a λ-Carrageenase Mutant with the Generation of Long-Chain λ-Neocarrageenan Oligosaccharides.Foods. 2024 Jun 18;13(12):1923. doi: 10.3390/foods13121923. Foods. 2024. PMID: 38928863 Free PMC article.
-
Biological pincer complexes.ChemCatChem. 2020 Sep 4;12(17):4242-4254. doi: 10.1002/cctc.202000575. Epub 2020 May 8. ChemCatChem. 2020. PMID: 33072225 Free PMC article.
-
Multicopy expression of sigma factor RpoH reduces prodigiosin biosynthesis in Serratia marcescens FS14.Antonie Van Leeuwenhoek. 2023 Nov;116(11):1197-1208. doi: 10.1007/s10482-023-01875-4. Epub 2023 Sep 20. Antonie Van Leeuwenhoek. 2023. PMID: 37728826
References
-
- Duine J. A. (2001) Cofactor diversity in biological oxidations: implications and applications. Chem. Rec. 1, 74–83 - PubMed
-
- Westerling J., Frank J., and Duine J. A. (1979) The prosthetic group of methanol dehydrogenase from Hyphomicrobium X: electron spin resonance evidence for a quinone structure. Biochem. Biophys. Res. Commun. 87, 719–724 - PubMed
-
- Duine J. A., Frank J., and van Zeeland J. K. (1979) Glucose dehydrogenase from Acinetobacter calcoaceticus: a “quinoprotein.” FEBS Lett. 108, 443–446 - PubMed
-
- van Kleef M. A., and Duine J. A. (1988) Bacterial NAD(P)-independent quinate dehydrogenase is a quinoprotein. Arch. Microbiol. 150, 32–36 - PubMed
-
- Groen B. W., and Duine J. A. (1990) Quinoprotein alcohol dehydrogenase from Pseudomonas aeruginosa and quinohemoprotein alcohol dehydrogenase from Pseudomonas testosteroni. Methods Enzymol. 188, 33–39 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

