Histone acetyltransferases: challenges in targeting bi-substrate enzymes
- PMID: 27231488
- PMCID: PMC4881052
- DOI: 10.1186/s13148-016-0225-2
Histone acetyltransferases: challenges in targeting bi-substrate enzymes
Abstract
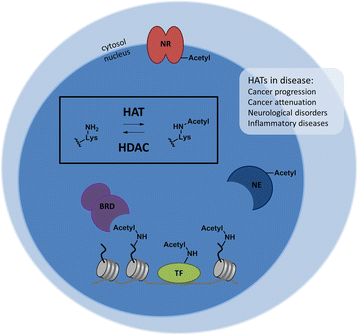
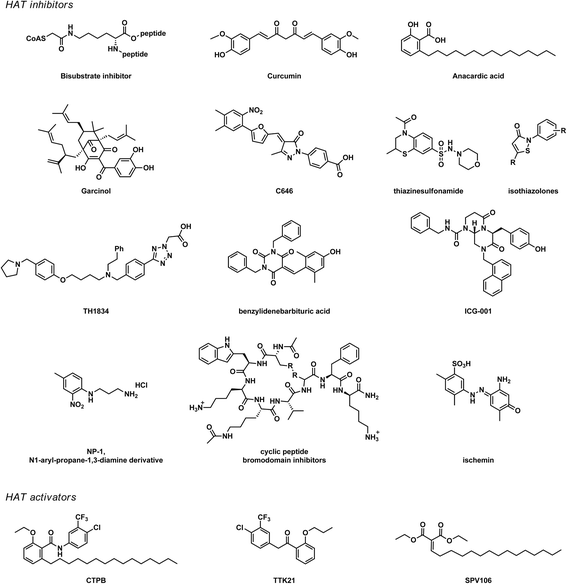
Histone acetyltransferases (HATs) are epigenetic enzymes that install acetyl groups onto lysine residues of cellular proteins such as histones, transcription factors, nuclear receptors, and enzymes. HATs have been shown to play a role in diseases ranging from cancer and inflammatory diseases to neurological disorders, both through acetylations of histone proteins and non-histone proteins. Several HAT inhibitors, like bi-substrate inhibitors, natural product derivatives, small molecules, and protein-protein interaction inhibitors, have been developed. Despite their potential, a large gap remains between the biological activity of inhibitors in in vitro studies and their potential use as therapeutic agents. To bridge this gap, new potent HAT inhibitors with improved properties need to be developed. However, several challenges have been encountered in the investigation of HATs and HAT inhibitors that hinder the development of new HAT inhibitors. HATs have been shown to function in complexes consisting of many proteins. These complexes play a role in the activity and target specificity of HATs, which limits the translation of in vitro to in vivo experiments. The current HAT inhibitors suffer from undesired properties like anti-oxidant activity, reactivity, instability, low potency, or lack of selectivity between HAT subtypes and other enzymes. A characteristic feature of HATs is that they are bi-substrate enzymes that catalyze reactions between two substrates: the cofactor acetyl coenzyme A (Ac-CoA) and a lysine-containing substrate. This has important-but frequently overlooked-consequences for the determination of the inhibitory potency of small molecule HAT inhibitors and the reproducibility of enzyme inhibition experiments. We envision that a careful characterization of molecular aspects of HATs and HAT inhibitors, such as the HAT catalytic mechanism and the enzyme kinetics of small molecule HAT inhibitors, will greatly improve the development of potent and selective HAT inhibitors and provide validated starting points for further development towards therapeutic agents.
Keywords: Catalytic mechanism; Epigenetics; HAT inhibitors; Histone acetyltransferases; Inhibitor kinetics; Lysine acetylation.
Figures

Similar articles
-
Histone acetyl transferases as emerging drug targets.Drug Discov Today. 2009 Oct;14(19-20):942-8. doi: 10.1016/j.drudis.2009.06.008. Epub 2009 Jul 2. Drug Discov Today. 2009. PMID: 19577000 Review.
-
Effective Quenchers Are Required to Eliminate the Interference of Substrate: Cofactor Binding in the HAT Scintillation Proximity Assay.Assay Drug Dev Technol. 2015 May;13(4):210-20. doi: 10.1089/adt.2015.636. Epub 2015 May 28. Assay Drug Dev Technol. 2015. PMID: 26065557 Free PMC article.
-
6-alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site.Eur J Med Chem. 2012 Jan;47(1):337-44. doi: 10.1016/j.ejmech.2011.11.001. Epub 2011 Nov 10. Eur J Med Chem. 2012. PMID: 22100137 Free PMC article.
-
Enzyme kinetics and inhibition of histone acetyltransferase KAT8.Eur J Med Chem. 2015 Nov 13;105:289-96. doi: 10.1016/j.ejmech.2015.10.016. Epub 2015 Oct 22. Eur J Med Chem. 2015. PMID: 26505788 Free PMC article.
-
Catalysis and substrate selection by histone/protein lysine acetyltransferases.Curr Opin Struct Biol. 2008 Dec;18(6):682-9. doi: 10.1016/j.sbi.2008.11.004. Curr Opin Struct Biol. 2008. PMID: 19056256 Free PMC article. Review.
Cited by
-
Protein acetylation and deacetylation: An important regulatory modification in gene transcription (Review).Exp Ther Med. 2020 Oct;20(4):2923-2940. doi: 10.3892/etm.2020.9073. Epub 2020 Jul 29. Exp Ther Med. 2020. PMID: 32855658 Free PMC article. Review.
-
A narrative review of the epigenetics of post-traumatic stress disorder and post-traumatic stress disorder treatment.Front Psychiatry. 2022 Nov 7;13:857087. doi: 10.3389/fpsyt.2022.857087. eCollection 2022. Front Psychiatry. 2022. PMID: 36419982 Free PMC article. Review.
-
HMGA1 Modulates Gene Transcription Sustaining a Tumor Signalling Pathway Acting on the Epigenetic Status of Triple-Negative Breast Cancer Cells.Cancers (Basel). 2019 Aug 2;11(8):1105. doi: 10.3390/cancers11081105. Cancers (Basel). 2019. PMID: 31382504 Free PMC article.
-
Intersection of Epigenetic and Metabolic Regulation of Histone Modifications in Acute Myeloid Leukemia.Front Oncol. 2019 May 22;9:432. doi: 10.3389/fonc.2019.00432. eCollection 2019. Front Oncol. 2019. PMID: 31192132 Free PMC article. Review.
-
Targeting epigenetic mechanisms as an emerging therapeutic strategy in pulmonary hypertension disease.Vasc Biol. 2020 Jan;2(1):R17-R34. doi: 10.1530/vb-19-0030. Epub 2020 Jan 9. Vasc Biol. 2020. PMID: 32161845 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

