SCF (Fbxl17) ubiquitylation of Sufu regulates Hedgehog signaling and medulloblastoma development
- PMID: 27234298
- PMCID: PMC4884786
- DOI: 10.15252/embj.201593374
SCF (Fbxl17) ubiquitylation of Sufu regulates Hedgehog signaling and medulloblastoma development
Abstract
Skp1-Cul1-F-box protein (SCF) ubiquitin ligases direct cell survival decisions by controlling protein ubiquitylation and degradation. Sufu (Suppressor of fused) is a central regulator of Hh (Hedgehog) signaling and acts as a tumor suppressor by maintaining the Gli (Glioma-associated oncogene homolog) transcription factors inactive. Although Sufu has a pivotal role in Hh signaling, the players involved in controlling Sufu levels and their role in tumor growth are unknown. Here, we show that Fbxl17 (F-box and leucine-rich repeat protein 17) targets Sufu for proteolysis in the nucleus. The ubiquitylation of Sufu, mediated by Fbxl17, allows the release of Gli1 from Sufu for proper Hh signal transduction. Depletion of Fbxl17 leads to defective Hh signaling associated with an impaired cancer cell proliferation and medulloblastoma tumor growth. Furthermore, we identify a mutation in Sufu, occurring in medulloblastoma of patients with Gorlin syndrome, which increases Sufu turnover through Fbxl17-mediated polyubiquitylation and leads to a sustained Hh signaling activation. In summary, our findings reveal Fbxl17 as a novel regulator of Hh pathway and highlight the perturbation of the Fbxl17-Sufu axis in the pathogenesis of medulloblastoma.
Keywords: Fbxl17; F‐box protein; Hedgehog signaling; Sufu; medulloblastoma.
© 2016 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

- A
Detection of Sufu and Skp1 after immunoprecipitation of the indicated Flag‐tagged F‐box proteins (FBPs). An empty vector (EV) was used as a negative control. HEK293T cells were treated with MLN4924 (2 μM) for 5 h prior to collection. Representative image of three independent experiments is shown.
- B, C
Immunoprecipitation of endogenous Sufu from PC3 (B) and DAOY cells (C) transfected with either an empty vector (EV) or Myc‐tagged Fbxl17. Nonspecific rabbit immunoglobulin G (IgG) was used as a negative control. Detection of light chains of immunoglobulin (IgG) was used to assess the amount of IgG used for each immunoprecipitation reaction. Treatment with MLN4925 (2 μM) was started 5 h before the cell collection.
- D
Quantification of Fbxl17 mRNA levels in PC3 and DAOY cells transfected with a nontargeting siRNA (Control) or two siRNAs to Fbxl17 (1) and (2). The cells were serum‐starved for 24 h in serum‐reduced medium and treated with SAG (100 nM) for 24 h prior to collection. Data are shown as mean ± SEM.
- E
Sufu protein levels in PC3 and DAOY cells treated as in (D). GAPDH was used as a loading control. Representative image of three independent experiments is shown.
- F
Sufu protein levels in PC3 and DAOY cells infected with an empty backbone retrovirus (EV) or a retrovirus expressing Myc‐tagged Fbxl17. Representative image from three independent experiments is shown.
- G
Detection of Sufu protein levels following cycloheximide (CHX) treatment for the indicated hours, and Fbxl17 depletion using two different siRNAs (Hrs = h). Representative image of two independent experiments is shown.
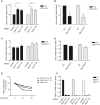
Relative abundance of Sufu protein levels in PC3 and DAOY cells reported in Fig 1E (mean ± SEM from three independent experiments, *P < 0.05; **P < 0.005).
Relative abundance of Sufu protein levels in PC3 and DAOY cells reported in Fig 1F (mean ± SEM from three independent experiments, **P < 0.005).
Sufu mRNA levels in PC3 and DAOY cells transfected with either a nontargeting siRNA (Control) or two siRNAs to Fbxl17 (1) and (2) (mean ± SEM).
Sufu mRNA levels after the transfection of the indicated cell lines with an empty vector (EV) or a vector expressing Myc‐Fbxl17 (mean ± SEM).
Relative abundance of Sufu protein levels in HeLa cells reported Fig 1G. Data are normalized relative to 0 h (Hrs) of CHX (cycloheximide) treatment.
Fbxl17 mRNA levels in HeLa cells treated as in Fig 1G (mean ± SEM).
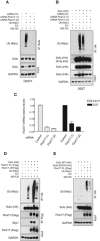
Detection of polyubiquitylated species of endogenous Sufu co‐immunopurified from DAOY cells transfected with Myc‐tagged ubiquitin (Ub) either in the presence of nontargeting siRNA (Control) or in the presence of two siRNAs against Fbxl17 (1) and (2). MG132 (10 μM) was added in all samples.
Detection of polyubiquitylated species of Sufu upon co‐transfection of HEK293T cells with HA‐tagged Sufu and Myc‐tagged ubiquitin (Ub) either in the presence of nontargeting siRNA (Control) or in the presence of two siRNAs against Fbxl17 (1) and (2). MG132 (10 μM) was added in all samples.
Fbxl17 mRNA relative levels in DAOY and HEK293T cells upon Fbxl17 depletion with either a nontargeting siRNA (Control) or two siRNAs to Fbxl17 (1) and (2). Data are shown as mean ± SEM.
Detection of ubiquitylated Sufu co‐immunoprecipitated from HEK293T cells co‐transfected with HA‐tagged Sufu, Myc‐ubiquitin (Ub) along with either Flag‐tagged Fbxl17 wild type (WT) or a mutant lacking the F‐box domain (Fbxl17ΔF). MG132 (10 μM) was added in all samples.
Detection of ubiquitylated Sufu co‐immunoprecipitated from HEK293T cells co‐transfected with Myc‐tagged ubiquitin (Ub), Flag‐tagged Fbxl17, and HA‐tagged Sufu WT or Sufu mutant K257R. MG132 (10 μM) was added in all samples.
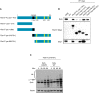
Scheme representing the mapping strategy to identify Fbxl17 region required for Sufu recognition. The F‐box is highlighted in black and the leucine‐rich repeats (LRR) in yellow.
Detection of Sufu and Skp1 after immunoprecipitation of Myc‐tagged Fbxl17‐truncated fragments, as indicated. An empty vector (EV) was used as a negative control. HEK293T cells were treated with MLN4924 (2 μM) for 5 h prior to collection.
In vitro ubiquitylation assay of Sufu WT and Sufu mutant K257R by Fbxl17‐truncated version 318–701. All proteins were synthesized in vitro using a T7‐coupled reticulocyte lysate system and incubated for the indicated times. Arrow indicates a nonspecific band detected in reticulocyte lysates.

Schematic representation of Sufu‐truncated mutants used to identify Sufu region required for Fbxl17 recognition. The black square delimitates the minimal binding region 350–425.
Detection of Myc‐tagged Fbxl17 after immunoprecipitation of HA‐Sufu‐truncated fragments, as indicated in (A). An empty vector (EV) was used as a negative control. HEK293T cells were treated with MLN4924 (2 μM) for 5 h prior to collection.
Detection of Myc‐tagged Fbxl17 binding to HA‐tagged Sufu deletion construct 350–425 in HEK293T cells. Transfection of an empty vector (EV) was used as a negative control for immunoprecipitation. Cells were treated with MLN4924 (2 μM) for 5 h prior to collection.
Detection of Myc‐tagged Fbxl17 binding to immunoprecipitated HA‐tagged Sufu wild type (WT), Sufu S352F, or Sufu S352D. An empty vector (EV) was used as a negative control. HEK293T cells were treated with MLN4924 (2 μM) for 5 h prior to collection.
Detection of Flag‐tagged Gli1 and Myc‐tagged Fbxl17 after immunoprecipitation of HA‐tagged Sufu WT or Sufu S342/6A and S342/6D, as indicated. HEK293T cells were treated with MLN4924 (2 μM) for 5 h prior to collection.
Detection of Myc‐tagged Fbxl17 after immunoprecipitation of the indicated HA‐Sufu point mutants in HEK293T cells. Cells were treated with MLN4924 (2 μM) for 5 h prior to collection.

Detection of Myc‐tagged Fbxl17 after immunoprecipitation of HA‐tagged Sufu WT or Sufu S342/6A and S342/6D, as indicated. HEK293T cells were treated with MLN4924 (2 μM) for 5 h prior to collection.
Detection of Flag‐tagged Gli1 and Myc‐tagged Fbxl17 binding to immunoprecipitated HA‐tagged Sufu wild type (WT) or to Sufu S352F. An empty vector (EV) was used as a negative control. HEK293T cells were treated with MLN4924 (2 μM) for 5 h prior to collection.
Detection of Sufu phosphorylated on S352/T353 after immunoprecipitation of HA‐tagged Sufu WT and Sufu 351–353 AAA, as indicated.
Detection of Myc‐tagged Fbxl17 binding to Sufu full‐length (FL) or to the following Sufu peptides: Sufu 340–360 (APSRKDSLESDSSTAIIPHEL); Sufu 340–360 [S352P/T353P] (phosphorylated on the residues S352 and T353); Sufu 351–372 (SSTAIIPHELIRTRQLESVHLK).
Detection of HA‐tagged Sufu binding to a Myc‐tagged Fbxl17 construct encompassing the residues 318–701 (corresponding to the F‐box motif and C‐terminus region) assessed by in vitro binding assay. Both proteins were synthesized in vitro using a T7‐coupled reticulocyte lysate system.
Detection of ubiquitylated Sufu immunoprecipitated from HEK293T cells co‐transfected with HA‐tagged ubiquitin (Ub), Myc‐tagged Fbxl17, and Flag‐tagged Gli1. MG132 (10 μM) was added in all samples.
Detection of Flag‐tagged Gli1 binding to Myc‐tagged Fbxl17 co‐immunoprecipitated from HEK293T cells transfected with a nontargeting siRNA (Control) or two siRNA to Sufu (1) and (2), as indicated.

Sufu protein levels in mouse embryonic fibroblasts (MEFs) Ptch1 +/+, Ptch1 +/−, and Ptch1 −/− transfected with a nontargeting siRNA (Control) or two siRNAs targeting mouse Fbxl17 (1) and (2).
Quantification of Fbxl17 mRNA levels in MEFs Ptch1 +/+, Ptch1 +/−, and Ptch1 −/− transfected as in (A). Data are shown as mean ± SEM.
Analysis of Gli1 mRNA levels in MEFs Ptch1 +/+ and Ptch1 −/− upon Fbxl17 depletion using two different siRNAs (mean ± SEM from three independent experiments, ***P < 0.0005, unpaired t‐test).
Analysis of Bcl2 mRNA levels in MEFs Ptch1 +/+ and Ptch1 −/− upon Fbxl17 depletion using two different siRNAs (mean ± SEM from three independent experiments, **P < 0.005, ***P < 0.0005, unpaired t‐test).
Detection of Flag‐tagged Gli1 binding to HA‐tagged Sufu in MEFs Ptch1 −/− upon expression of Myc‐tagged Fbxl17, or Fbxl17 depletion by two siRNAs. For each condition, 200 μg of whole‐cell lysate was immunoprecipitated.
Immunostaining of Fbxl17 using anti‐Myc antibody in Ptch1 −/− MEFs stably transduced with a pBABE vector expressing Fbxl17 tagged with Myc either at the N‐terminus [Myc(N)‐Fbxl17] or at the C‐terminus [Myc(C)‐Fbxl17]. Scale bars: 20 μm.
Detection of Fbxl17 using anti‐Myc antibody in Ptch1 −/− MEFs stably expressing Myc(N)‐Fbxl17 (Fbxl17 tagged at the N‐terminus) and Myc(C)‐Fbxl17 (Fbxl17 tagged at the C‐terminus).
Detection of HA‐tagged Fbxl17 binding to endogenous Sufu immunoprecipitated from cytoplasmic and nuclear extracts from Ptch1 −/− MEFs. Asterisk (*) indicates a nonspecific band. Identification of cytoplasmic and nuclear fractions was performed by lamin A/C and GAPDH detection.
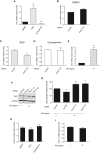
Gli1 mRNA levels in DAOY cells upon the treatment with SAG or cyclopamine. DMSO was used as a control vehicle (mean ± SEM from three independent experiments, **P < 0.005; ***P < 0.0005).
Gli1 mRNA levels in DAOY cells transfected with nontargeting siRNA (Control) or siRNAs against Fbxl17 (2) and treated with DMSO (mean ± SEM).
Gli1 mRNA levels in DAOY cells transfected as in (B) and treated with SAG (mean ± SEM from three independent experiments, *P < 0.05).
Gli1 mRNA levels in DAOY cells transfected as in (B) and treated with cyclopamine (mean ± SEM).
Gli1 mRNA levels in DAOY cells treated with control medium (−) or medium containing SHH ligand (+) (mean ± SEM from three independent experiments, **P < 0.005).
Detection of Sufu protein levels in DAOY in the presence of a nontargeting siRNA (Control) or two siRNAs to Fbxl17 (1) and (2), after the treatment with control medium (−) or medium containing SHH (Sonic Hedgehog) ligand (+).
Relative abundance of Sufu protein levels detected in (F) (mean ± SEM from three independent experiments, **P < 0.005; ***P < 0.05).
Fbxl17 mRNA levels in DAOY cells upon the treatment with DMSO, SAG, or cyclopamine (mean ± SEM).
Fbxl17 mRNA levels in DAOY cells after the treatment with control medium (−) or medium containing SHH ligand (+) (mean ± SEM).
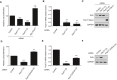
Cell proliferation of DAOY cells upon Fbxl17 depletion using a nontargeting siRNA (Control), two siRNAs against Fbxl17 (1) and (2) or upon reintroduction of a Myc‐tagged Fbxl17 construct in Fbxl17‐depleted DAOY cells using siRNA Fbxl17 (2) (mean ± SEM from three independent experiments, **P < 0.005; ***P < 0.0005, unpaired t‐test).
Quantification of Fbxl17 mRNA levels in DAOY cells transfected with a nontargeting siRNA (Control) or two siRNAs against Fbxl17 (1) and (2) (mean ± SEM from three independent experiments, **P < 0.005, unpaired t‐test).
Sufu protein levels in DAOY cells treated as in (A). Representative image of three independent experiments is shown.
Cell proliferation of DAOY cells transfected with nontargeting siRNA (Control), siRNA against Fbxl17 or a combination of siRNA targeting Fbxl17 and Sufu (mean ± SEM from three independent experiments, **P < 0.005; ***P < 0.0005, unpaired t‐test).
Quantification of Fbxl17 mRNA levels in DAOY cells treated as in (D) (mean ± SEM from three independent experiments, ***P < 0.0005, unpaired t‐test).
Sufu protein levels in DAOY cells treated as in (D). Representative image of three independent experiments is shown.
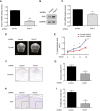
Quantification of Fbxl17 mRNA levels in DAOY cells transfected with control shRNA or shRNA targeting Fbxl17 (mean ± SEM from three independent experiments, ***P < 0.0005, unpaired t‐test).
Detection of Sufu protein levels in DAOY cells transfected with control shRNA or shRNA Fbxl17. Representative image of three independent experiments is shown.
Quantification of Gli1 mRNA levels in DAOY cells transfected with control shRNA or shRNA Fbxl17 (mean ± SEM from three independent experiments, **P < 0.005, unpaired t‐test).
T2‐weighted magnetic resonance images showing tumor development in rats injected with DAOY cells transfected with control shRNA or shRNA against Fbxl17. One representative image for each condition at 10 weeks is shown. Scale bar: 5 mm.
Graph showing area of T2 hyperintensity (a surrogate marker of tumor growth) between 4 and 10 weeks post‐tumor induction. Animals were injected with DAOY cells stably expressing control shRNA or shRNA against Fbxl17 (10,000 cells/μl) (mean ± SEM; n = 5; two‐way ANOVA, followed by unpaired t‐test, *P < 0.05, **P < 0.01).
Representative immunohistochemical image of vimentin staining. Scale bar: 1 mm.
Quantification of tumor growth (vimentin staining) in rats injected with DAOY cells transfected with either control shRNA or shRNA against Fbxl17 (mean ± SEM; n = 5; unpaired t‐test, *P < 0.05).
Representative immunohistochemical image of Ki67 staining. Scale bar: 0.5 mm/0.1 mm.
Quantification of proliferative index (Ki67 staining) in rats injected with DAOY cells transfected with either control shRNA or shRNA against Fbxl17 (mean ± SEM; n = 5; unpaired t‐test, *P < 0.05).
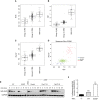
- A–C
mRNA expression of Sufu, Gli1, and Fbxl17 detected in 285 medulloblastoma samples (GEO accession: GSE37382), stratified by tumor subtype. Level of significance (P) for Kruskal–Wallis (KW) rank test is P < 10−8. Analysis using one‐way ANOVA gave comparable results (median ± 1.5 interquartile distance).
- D
Expression of Gli1 and Fbxl17 in the 285 patients is shown colored by subgroup; level of significance (P) for Kruskal–Wallis (KW) rank test is P < 10−8, and Spearman's rank correlation (R = 0.5640) is indicated.
- E
Detection of HA‐tagged Sufu wild type (WT) and HA‐tagged Sufu S352F protein levels in NIH3T3 cells after cycloheximide (CHX) treatment. Where indicated, the cells were transfected with a nontargeting siRNA (Control) or two siRNA to Fbxl17 (1) and (2). Representative image of three independent experiments is shown.
- F
Assessment of Hh pathway activation using Gli1‐luciferase reporter assay in MEFs Sufu −/− upon the reintroduction of HA‐tagged Sufu WT or Sufu mutant S352F using a retroviral system (mean ± SEM from three independent experiments, **P < 0.005; ***P < 0.0005, unpaired t‐test).

Fbxl17 mRNA levels in GCPs after transfection with an empty vector (pCDNA) or a vector expressing Myc‐Fbxl17 (Fbxl17) (mean ± SEM from three independent experiments, **P < 0.005).
Cell proliferation of GCPs upon Fbxl17 overexpression (mean ± SEM from three independent experiments, **P < 0.005).
Fbxl17 mRNA levels in GCPs transfected with either a nontargeting siRNA (Control) or siRNAs to Fbxl17 (mean ± SEM from three independent experiments, **P < 0.005).
Cell proliferation of GCPs upon Fbxl17 depletion (mean ± SEM from four independent experiments, **P < 0.005).
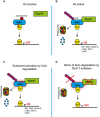
- A–D
Scheme outlining the mechanism of Sufu recognition by Fbxl17 and its alteration in medulloblastoma.
References
-
- Bhatia N, Thiyagarajan S, Elcheva I, Saleem M, Dlugosz A, Mukhtar H, Spiegelman VS (2006) Gli2 is targeted for ubiquitination and degradation by beta‐TrCP ubiquitin ligase. J Biol Chem 281: 19320–19326 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

