Genomic and transcriptomic predictors of response levels to endurance exercise training
- PMID: 27234805
- PMCID: PMC5407970
- DOI: 10.1113/JP272559
Genomic and transcriptomic predictors of response levels to endurance exercise training
Abstract
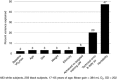
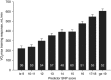
Predicting the responsiveness to regular exercise is a topic of great relevance due to its potential role in personalized exercise medicine applications. The present review focuses on cardiorespiratory fitness (commonly measured by maximal oxygen uptake, V̇O2 max ), a trait with wide-ranging impact on health and performance indicators. Gains in V̇O2 max demonstrate large inter-individual variation even in response to standardized exercise training programmes. The estimated ΔVO2 max heritability of 47% suggests that genomic-based predictors alone are insufficient to account for the total trainability variance. Candidate gene and genome-wide linkage studies have not significantly contributed to our understanding of the molecular basis of trainability. A genome-wide association study suggested that V̇O2 max trainability is influenced by multiple genes of small effects, but these findings still await rigorous replication. Valuable evidence, however, has been obtained by combining skeletal muscle transcript abundance profiles with common DNA variants for the prediction of the V̇O2 max response to exercise training. Although the physiological determinants of V̇O2 max measured at a given time are largely enunciated, what is poorly understood are the details of tissue-specific molecular mechanisms that limit V̇O2 max and related signalling pathways in response to exercise training. Bioinformatics explorations based on thousands of variants have been used to interrogate pathways and systems instead of single variants and genes, and the main findings, along with those from exercise experimental studies, have been summarized here in a working model of V̇O2 max trainability.
© 2016 The Authors. The Journal of Physiology © 2016 The Physiological Society.
Figures
References
-
- Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent LC, De Moor B, Marynen P, Hassan B, Carmeliet P & Moreau Y (2006). Gene prioritization through genomic data fusion. Nat Biotechnol 24, 537–544. - PubMed
-
- Bennett AF (1987). Interindividual variability: an underutilized resource In New Directions in Ecological Physiology, ed. Feder ME, Bennett A, Burggren W. & Huey R, pp. 147–169. Cambridge University Press, Cambridge, New York.
-
- Blache D, Lussier‐Cacan S, Gagnon J, Leon AS, Rao DC, Skinner JS, Wilmore JH, Rankinen T, Bouchard C & Davignon J (2007). Effect of exercise training on in vitro LDL oxidation and free radical‐induced hemolysis: the HERITAGE Family Study. Antioxid Redox Signal 9, 123–130. - PubMed
-
- Blomqvist CG & Saltin B (1983). Cardiovascular adaptations to physical training. Annu Rev Physiol 45, 169–189. - PubMed
-
- Bouchard C (1983). Human adaptability may have a genetic basis In Health Risk Estimation, Risk Reduction and Health Promotion. Proceedings of the 18th Annual Meeting of the Society of Prospective Medicine, ed. Landry F, pp. 463–476. Canadian Public Health Association, Ottawa.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

