Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing
- PMID: 27237775
- PMCID: PMC5752108
- DOI: 10.1016/j.mimet.2016.05.022
Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing
Abstract
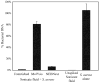
Metagenomic whole genome sequencing for detection of pathogens in clinical samples is an exciting new area for discovery and clinical testing. A major barrier to this approach is the overwhelming ratio of human to pathogen DNA in samples with low pathogen abundance, which is typical of most clinical specimens. Microbial DNA enrichment methods offer the potential to relieve this limitation by improving this ratio. Two commercially available enrichment kits, the NEBNext Microbiome DNA Enrichment Kit and the Molzym MolYsis Basic kit, were tested for their ability to enrich for microbial DNA from resected arthroplasty component sonicate fluids from prosthetic joint infections or uninfected sonicate fluids spiked with Staphylococcus aureus. Using spiked uninfected sonicate fluid there was a 6-fold enrichment of bacterial DNA with the NEBNext kit and 76-fold enrichment with the MolYsis kit. Metagenomic whole genome sequencing of sonicate fluid revealed 13- to 85-fold enrichment of bacterial DNA using the NEBNext enrichment kit. The MolYsis approach achieved 481- to 9580-fold enrichment, resulting in 7 to 59% of sequencing reads being from the pathogens known to be present in the samples. These results demonstrate the usefulness of these tools when testing clinical samples with low microbial burden using next generation sequencing.
Keywords: Clinical samples; Enrichment; Metagenomics; Pathogen detection; Whole genome sequencing.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures
References
-
- Antequera F, Tamame M, Villanueva JR, Santos T. DNA methylation in the fungi. J Biol Chem. 1984;259:8033–8036. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

