Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes
- PMID: 27238021
- PMCID: PMC4912438
- DOI: 10.1016/j.cell.2016.05.007
Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes
Abstract
Understanding how neural information is processed in physiological and pathological states would benefit from precise detection, localization, and quantification of the activity of all neurons across the entire brain, which has not, to date, been achieved in the mammalian brain. We introduce a pipeline for high-speed acquisition of brain activity at cellular resolution through profiling immediate early gene expression using immunostaining and light-sheet fluorescence imaging, followed by automated mapping and analysis of activity by an open-source software program we term ClearMap. We validate the pipeline first by analysis of brain regions activated in response to haloperidol. Next, we report new cortical regions downstream of whisker-evoked sensory processing during active exploration. Last, we combine activity mapping with axon tracing to uncover new brain regions differentially activated during parenting behavior. This pipeline is widely applicable to different experimental paradigms, including animal species for which transgenic activity reporters are not readily available.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
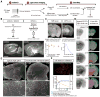

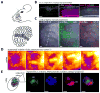



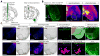
Comment in
-
Catching the Brain in the Act.Cell. 2016 Jun 16;165(7):1570-1571. doi: 10.1016/j.cell.2016.06.008. Cell. 2016. PMID: 27315474
References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B … 1995
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

