A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria
- PMID: 27238023
- PMCID: PMC4894308
- DOI: 10.1016/j.cell.2016.05.003
A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria
Abstract
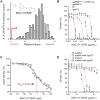
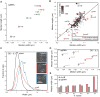
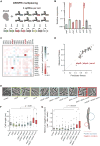
Essential gene functions underpin the core reactions required for cell viability, but their contributions and relationships are poorly studied in vivo. Using CRISPR interference, we created knockdowns of every essential gene in Bacillus subtilis and probed their phenotypes. Our high-confidence essential gene network, established using chemical genomics, showed extensive interconnections among distantly related processes and identified modes of action for uncharacterized antibiotics. Importantly, mild knockdown of essential gene functions significantly reduced stationary-phase survival without affecting maximal growth rate, suggesting that essential protein levels are set to maximize outgrowth from stationary phase. Finally, high-throughput microscopy indicated that cell morphology is relatively insensitive to mild knockdown but profoundly affected by depletion of gene function, revealing intimate connections between cell growth and shape. Our results provide a framework for systematic investigation of essential gene functions in vivo broadly applicable to diverse microorganisms and amenable to comparative analysis.
Published by Elsevier Inc.
Figures




References
-
- Blomen VA, Májek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

