NMR-based platform for fragment-based lead discovery used in screening BRD4-targeted compounds
- PMID: 27238211
- PMCID: PMC4933754
- DOI: 10.1038/aps.2016.19
NMR-based platform for fragment-based lead discovery used in screening BRD4-targeted compounds
Abstract
Aim: Fragment-based lead discovery (FBLD) is a complementary approach in drug research and development. In this study, we established an NMR-based FBLD platform that was used to screen novel scaffolds targeting human bromodomain of BRD4, and investigated the binding interactions between hit compounds and the target protein.
Methods: 1D NMR techniques were primarily used to generate the fragment library and to screen compounds. The inhibitory activity of hits on the first bromodomain of BRD4 [BRD4(I)] was examined using fluorescence anisotropy binding assay. 2D NMR and X-ray crystallography were applied to characterize the binding interactions between hit compounds and the target protein.
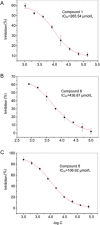
Results: An NMR-based fragment library containing 539 compounds was established, which were clustered into 56 groups (8-10 compounds in each group). Eight hits with new scaffolds were found to inhibit BRD4(I). Four out of the 8 hits (compounds 1, 2, 8 and 9) had IC50 values of 100-260 μmol/L, demonstrating their potential for further BRD4-targeted hit-to-lead optimization. Analysis of the binding interactions revealed that compounds 1 and 2 shared a common quinazolin core structure and bound to BRD4(I) in a non-acetylated lysine mimetic mode.
Conclusion: An NMR-based platform for FBLD was established and used in discovery of BRD4-targeted compounds. Four potential hit-to-lead optimization candidates have been found, two of them bound to BRD4(I) in a non-acetylated lysine mimetic mode, being selective BRD4(I) inhibitors.
Figures


Similar articles
-
Straightforward hit identification approach in fragment-based discovery of bromodomain-containing protein 4 (BRD4) inhibitors.Bioorg Med Chem. 2018 Jul 23;26(12):3399-3405. doi: 10.1016/j.bmc.2018.05.010. Epub 2018 May 9. Bioorg Med Chem. 2018. PMID: 29764756
-
Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking.Bioorg Med Chem Lett. 2014 Jun 1;24(11):2493-6. doi: 10.1016/j.bmcl.2014.04.017. Epub 2014 Apr 13. Bioorg Med Chem Lett. 2014. PMID: 24767840
-
Cell-based protein stabilization assays for the detection of interactions between small-molecule inhibitors and BRD4.J Biomol Screen. 2015 Feb;20(2):180-9. doi: 10.1177/1087057114552398. Epub 2014 Sep 29. J Biomol Screen. 2015. PMID: 25266565
-
Process of Fragment-Based Lead Discovery-A Perspective from NMR.Molecules. 2016 Jul 16;21(7):854. doi: 10.3390/molecules21070854. Molecules. 2016. PMID: 27438813 Free PMC article. Review.
-
Combining NMR and X-ray crystallography in fragment-based drug discovery: discovery of highly potent and selective BACE-1 inhibitors.Top Curr Chem. 2012;317:83-114. doi: 10.1007/128_2011_183. Top Curr Chem. 2012. PMID: 21647837 Review.
Cited by
-
A simple fluorescent assay for the discovery of protein-protein interaction inhibitors.Anal Biochem. 2019 Mar 15;569:46-52. doi: 10.1016/j.ab.2019.01.010. Epub 2019 Jan 30. Anal Biochem. 2019. PMID: 30707898 Free PMC article.
-
Applications of Solution NMR in Drug Discovery.Molecules. 2021 Jan 22;26(3):576. doi: 10.3390/molecules26030576. Molecules. 2021. PMID: 33499337 Free PMC article. Review.
-
Paramagnetic relaxation enhancement for protein-observed 19F NMR as an enabling approach for efficient fragment screening.RSC Adv. 2016;6(98):95715-95721. doi: 10.1039/C6RA21226C. Epub 2016 Sep 29. RSC Adv. 2016. PMID: 28496971 Free PMC article.
-
Alternative Mechanisms for DNA Engagement by BET Bromodomain-Containing Proteins.Biochemistry. 2022 Jul 5;61(13):1260-1272. doi: 10.1021/acs.biochem.2c00157. Epub 2022 Jun 24. Biochemistry. 2022. PMID: 35748495 Free PMC article.
-
Allosteric Regulation of Hsp90α's Activity by Small Molecules Targeting the Middle Domain of the Chaperone.iScience. 2020 Feb 21;23(2):100857. doi: 10.1016/j.isci.2020.100857. Epub 2020 Jan 21. iScience. 2020. PMID: 32058968 Free PMC article.
References
-
- Erlanson DA. Introduction to fragment-based drug discovery. Top Curr Chem 2012; 317: 1–32. - PubMed
-
- Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J Med Chem 2008; 51: 3661–80. - PubMed
-
- Hanzawa H, Takizawa T. NMR in the fragment-based drug discovery. Tanpakushitsu Kakusan Koso 2009; 54 (12 Suppl): 1617–21. Japanese. - PubMed
-
- Campos-Olivas R. NMR screening and hit validation in fragment based drug discovery. Curr Top Med Chem 2011; 11: 43–67. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

